SpoIIAB
- Description: anti-SigF/ protein serine kinase
| Gene name | spoIIAB |
| Synonyms | |
| Essential | no |
| Product | anti-SigF/ protein serine kinase |
| Function | control of SigF activity septation; phosphorylation and inactivation of SpoIIAA |
| Gene expression levels in SubtiExpress: spoIIAB | |
| Interactions involving this protein in SubtInteract: SpoIIAB | |
| Metabolic function and regulation of this protein in SubtiPathways: Stress | |
| MW, pI | 16 kDa, 4.383 |
| Gene length, protein length | 438 bp, 146 aa |
| Immediate neighbours | sigF, spoIIAA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 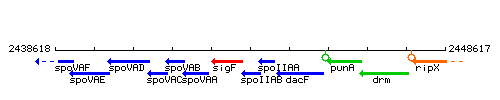
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed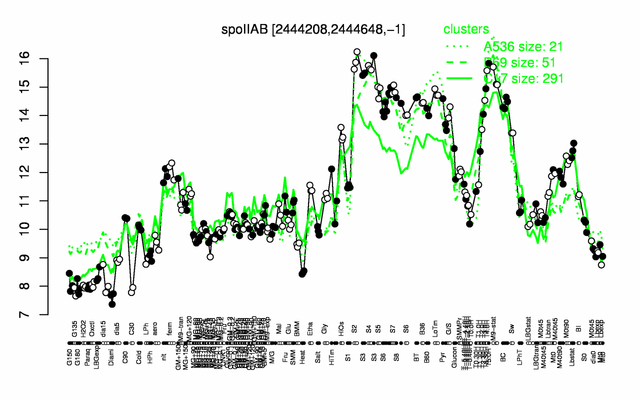
| |
Contents
Categories containing this gene/protein
protein modification, sigma factors and their control, sporulation proteins
This gene is a member of the following regulons
AbrB regulon, SigF regulon, SigG regulon, SigH regulon, SinR regulon, Spo0A regulon
The gene
Basic information
- Locus tag: BSU23460
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + a protein = ADP + a phosphoprotein (according to Swiss-Prot)
- Protein family: anti-sigma-factor family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure: 1L0O (complex with SigF, Geobacillus stearothermophilus), 1TIL (complex with SpoIIAA, Geobacillus stearothermophilus)
- UniProt: P10728
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Charles Moran, Emory University, NC, USA homepage
Your additional remarks
References
Modeling of SigF activation
Original Publications