Difference between revisions of "RocG"
(→The gene) |
|||
| Line 29: | Line 29: | ||
__TOC__ | __TOC__ | ||
| − | + | <br/><br/> | |
Revision as of 22:39, 13 January 2009
- Description: catabolic glutamate dehydrogenase induced by arginine, ornithine or proline, subject to carbon catabolite repression
| Gene name | rocG |
| Synonyms | |
| Essential | no |
| Product | glutamate dehydrogenase (major) |
| Function | arginine utilization, controls the activity of GltC |
| MW, pI | 46.2 kDa, 6.28 |
| Gene length, protein length | 1272 bp, 424 amino acids |
| Immediate neighbours | yweA, rocA |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 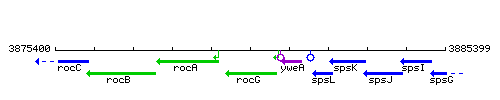
| |
Contents
The gene
Basic information
- Coordinates: 3879765 - 3881036
Phenotypes of a mutant
Poor growth on complex media such as LB. No growth in minimal media with arginine as the only carbon source. Rapid accumulation of suppressor mutants (gudB1)
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: L-glutamate + H(2)O + NAD(+) = 2-oxoglutarate + NH(3) + NADH
- Protein family: Glu/Leu/Phe/Val dehydrogenases family
- Paralogous protein(s): GudB
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions: RocG-GltC, this interaction prevents transcription activation of the gltAB operon by GltC PubMed
- Localization:
Database entries
- Structure:
- Swiss prot entry: [3]
- KEGG entry: [4]
- E.C. number: [5]
Additional information
Expression and regulation
- Operon: rocG
- Regulation: induced by arginine (RocR, AhrC), ornithine or proline, subject to carbon catabolite repression (CcpA)
- Regulatory mechanism: RocR: transcription activation PubMedPubMed; AhrC: transcription activation ; CcpA: transcription repression
- Additional information:
Activation by RocR requires binding of RocG to a downstream element PubMed
Biological materials
Labs working on this gene/protein
Linc Sonenshein, Tufts University, Boston, MA, USA Homepage
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References
- Commichau, F. M., Wacker, I., Schleider, J., Blencke, H.-M., Reif, I., Tripal, P., and Stülke, J. (2007) Characterization of Bacillus subtilis mutants with carbon source-independent glutamate biosynthesis. J Mol Microbiol Biotechnol 12: 106-113. PubMed
- Commichau, F. M., Herzberg, C., Tripal, P., Valerius, O., and Stülke, J. (2007) A regulatory protein-protein interaction governs glutamate biosynthesis in Bacillus subtilis: The glutamate dehydrogenase RocG moonlights in controlling the transcription factor GltC. Mol Microbiol 65: 642-654. PubMed
- Commichau, F. M., Gunka, K., Landmann, J. J. & Stülke, J. (2008) Glutamate metabolism in Bacillus subtilis: Gene expression and enzyme activities evolved to avoid futile cycles and to allow rapid responses to perturbations in the system. J. Bacteriol. 190: 3557-3564. PubMed
- Herzberg, C., Flórez Weidinger, L. A., Dörrbecker, B., Hübner, S., Stülke, J. & Commichau, F. M. (2007) SPINE: A method for the rapid detection and analysis of protein-protein interactions in vivo. Proteomics 7: 4032-4035. PubMed
- Ali, N. O., J. Jeusset, E. Larquet, E. le Cam, B. Belitsky, A. L. Sonenshein, T. Msadek, and M. Débarbouillé. 2003. Specificity of the interaction of RocR with the rocG-rocA intergenic region in Bacillus subtilis. Microbiology 149: 739-750. PubMed
- Belitsky BR, Sonenshein AL (1998) Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J Bacteriol 180:6298-6305 PubMed
- Belitsky BR, Sonenshein, AL: An enhancer element located downstream of the major glutamate dehydrogenase gene of Bacillus subtilis. Proc Natl Acad Sci USA 1999, 96:10290-10295. PubMed
- Belitsky BR, Sonenshein, AL: CcpA-dependent regulation of Bacillus subtilis glutamate dehydrogenase gene expression. J Bacteriol 2004, 186:3392-3398. PubMed
- Belitsky BR, Sonenshein AL (2004) Modulation of activity of Bacillus subtilis regulatory proteins GltC and TnrA by glutamate dehydrogenase. J Bacteriol 186:3399-3407 PubMed
- Khan, M. I., K. Ito, H. Kim, H. Ashida, T. Ishikawa, H. Shibata, and Y. Sawa. 2005. Molecular properties and enhancement of thermostability by random mutagenesis of glutamate dehydrogenase from Bacillus subtilis. Biosci. Biotechnol. Biochem. 69: 1861-1870. PubMed
- Stillman TJ, Baker PJ, Britton KL, Rice DW Conformational flexibility in glutamate dehydrogenase. Role of water in substrate recognition and catalysis. J Mol Biol 1993, 234:1131-1139. PubMed