Difference between revisions of "Spo0A"
| Line 14: | Line 14: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || initiation of sporulation | |style="background:#ABCDEF;" align="center"|'''Function''' || initiation of sporulation | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/biofilm.html Biofilm], [http://subtiwiki.uni-goettingen.de/pathways/gene_regulation_nucleotides.html Nucleotides (regulation)], [http://subtiwiki.uni-goettingen.de/pathways/glutamate.html Ammonium/ glutamate], [http://subtiwiki.uni-goettingen.de/pathways/carbon_flow.html Central C-metabolism],<br/>[http://subtiwiki.uni-goettingen.de/pathways/carbohydrate_metabolic_pathways.html Sugar catabolism], [http://subtiwiki.uni-goettingen.de/pathways/stress_response.html Stress], [http://subtiwiki.uni-goettingen.de/pathways/tRNA_charging.html tRNA charging]''' | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/biofilm.html Biofilm], [http://subtiwiki.uni-goettingen.de/pathways/biofilm.html Biofilm], [http://subtiwiki.uni-goettingen.de/pathways/gene_regulation_nucleotides.html Nucleotides (regulation)], [http://subtiwiki.uni-goettingen.de/pathways/glutamate.html Ammonium/ glutamate], [http://subtiwiki.uni-goettingen.de/pathways/carbon_flow.html Central C-metabolism],<br/>[http://subtiwiki.uni-goettingen.de/pathways/carbohydrate_metabolic_pathways.html Sugar catabolism], [http://subtiwiki.uni-goettingen.de/pathways/stress_response.html Stress], [http://subtiwiki.uni-goettingen.de/pathways/tRNA_charging.html tRNA charging]''' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 29 kDa, 5.989 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 29 kDa, 5.989 | ||
Revision as of 13:59, 16 February 2010
- Description: phosphorelay regulator, initiation of sporulation
| Gene name | spo0A |
| Synonyms | spo0C, spo0G, spoIIL, sof-1 |
| Essential | no |
| Product | phosphorelay response regulator |
| Function | initiation of sporulation |
| Metabolic function and regulation of this protein in SubtiPathways: Biofilm, Biofilm, Nucleotides (regulation), Ammonium/ glutamate, Central C-metabolism, Sugar catabolism, Stress, tRNA charging | |
| MW, pI | 29 kDa, 5.989 |
| Gene length, protein length | 801 bp, 267 aa |
| Immediate neighbours | yqiG, spoIVB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 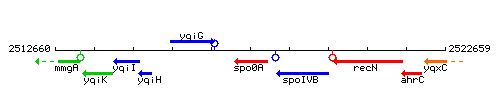
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU24220
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Genes controlled by Spo0A
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: receives phosphorylation from Spo0B, dephosphorylation by Spo0E, direct phosphorylation by KinC upon potassium leakage PubMed
- Cofactor(s):
- Effectors of protein activity: the phosphorylation state affects DNA binding activity
- Localization:
Database entries
- Structure: 1QMP (with phosphorylated aspartate, Geobacillus stearothermophilus), 1FC3 (trans-activation domain, Geobacillus stearothermophilus), 1LQ1 (complex with DNA)
- UniProt: P06534
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: spo0A PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Tony Wilkinson, York University, U.K. homepage
- Imrich Barak, Slovak Academy of Science, Bratislava, Slovakia homepage
- Charles Moran, Emory University, NC, USA homepage
Your additional remarks
References
Reviews
Daniel López, Roberto Kolter
Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis.
FEMS Microbiol Rev: 2010, 34(2);134-49
[PubMed:20030732]
[WorldCat.org]
[DOI]
(I p)
Daniel Schultz, Peter G Wolynes, Eshel Ben Jacob, José N Onuchic
Deciding fate in adverse times: sporulation and competence in Bacillus subtilis.
Proc Natl Acad Sci U S A: 2009, 106(50);21027-34
[PubMed:19995980]
[WorldCat.org]
[DOI]
(I p)
Original Publications