CitB
- Description: trigger enzyme: aconitase and RNA binding protein
| Gene name | citB |
| Synonyms | |
| Essential | no |
| Product | trigger enzyme: aconitate hydratase (aconitase) |
| Function | TCA cycle |
| Gene expression levels in SubtiExpress: citB | |
| Interactions involving this protein in SubtInteract: CitB | |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 99 kDa, 4.903 |
| Gene length, protein length | 2727 bp, 909 aa |
| Immediate neighbours | sspO, yneN |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 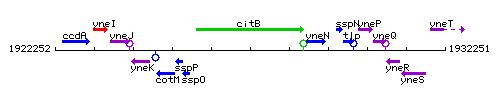
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed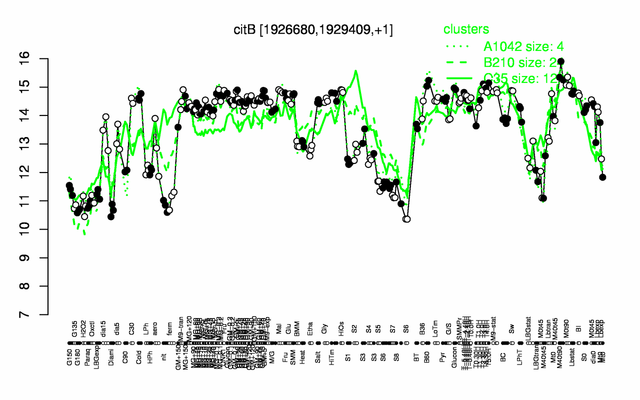
| |
Contents
Categories containing this gene/protein
carbon core metabolism, trigger enzyme, RNA binding regulators
This gene is a member of the following regulons
CcpA regulon, CcpC regulon, CodY regulon, FsrA regulon
The CitB regulon: feuA-feuB-feuC-ybbA, citZ
The gene
Basic information
- Locus tag: BSU18000
Phenotypes of a mutant
- glutamate auxotrophy and a defect in sporulation PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- A mutation was found in this gene after evolution under relaxed selection for sporulation PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Citrate <=> isocitrate
- Binding to iron responsive elements (IRE RNA) in the absence of the FeS cluster PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s): FeS cluster
- Effectors of protein activity:
Database entries
- Structure: 1L5J (E. coli)
- UniProt: P09339
- KEGG entry: [3]
- E.C. number: 4.2.1.3
Additional information
- B. subtilis aconitase is both an enzyme and an RNA binding protein (moonlighting protein) PubMed
- extensive information on the structure and enzymatic properties of CitB can be found at Proteopedia
Expression and regulation
- Regulation:
- repressed during growth in the presence of branched chain amino acids (CodY) PubMed
- repressed in the presence of glucose and glutamate (CcpC) PubMed
- expressed upon transition into the stationary phase (AbrB) PubMed, indirect negative regulation by AbrB PubMed
- repressed by glucose (3.7-fold) (CcpA) PubMed
- repression by glucose + arginine (CcpC) PubMed
- less expressed under conditions of extreme iron limitation (FsrA) PubMed
- part of the iron sparing response (FsrA) PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- GP683 (erm), available in Jörg Stülke's lab
- GP1441 (spc), available in Jörg Stülke's lab
- 1A999 ( citB::spec), PubMed, available at BGSC
- Expression vector:
- GP1439 (citB-Strep (spc)), purification from B. subtilis, for SPINE, available in Jörg Stülke's lab
- pGP1810 (for expression, purification in E. coli with N-terminal Strep-tag, in pGP172, available in Jörg Stülke's lab
- lacZ fusion:
- pGP700 (in pAC5), available in Jörg Stülke's lab
- GFP fusion: GP1434 (spc, based on pGP1870), available in Jörg Stülke's lab
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's lab
- Antibody: available in Linc Sonenshein's lab
- FLAG-tag construct:
- GP1144 (spc, based on pGP1331), available in Jörg Stülke's lab
- GP1145 (kan), available in Jörg Stülke's lab
Labs working on this gene/protein
- Linc Sonenshein, Tufts University, Boston, MA, USA Homepage
- Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References
Reviews
Original publications