FrlB
- Description: fructoselysine-6-P-glycosidase
| Gene name | frlB |
| Synonyms | yurP |
| Essential | no |
| Product | fructoselysine-6-P-glycosidase |
| Function | metabolism of aminoacylated fructose |
| Gene expression levels in SubtiExpress: frlB | |
| Metabolic function and regulation of this protein in SubtiPathways: Alternative nitrogen sources | |
| MW, pI | 36 kDa, 5.442 |
| Gene length, protein length | 984 bp, 328 aa |
| Immediate neighbours | frlO, yurQ |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 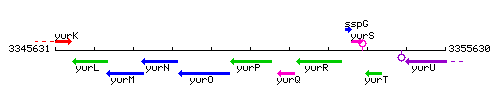
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed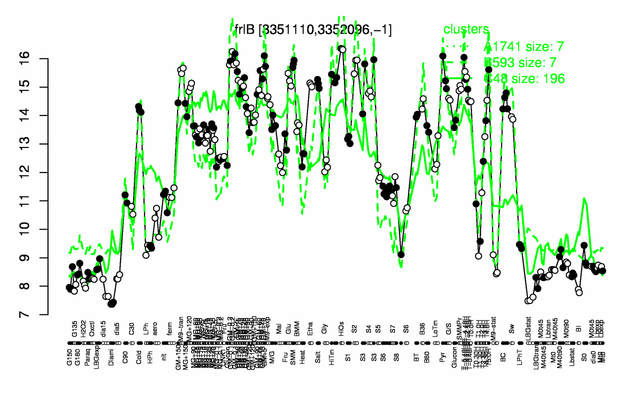
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources, utilization of nitrogen sources other than amino acids, membrane proteins, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU32610
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- phosphorylated on Arg-48 PubMed
- Cofactor(s):
- Effectors of protein activity:
- Localization: membrane associated PubMed
Database entries
- Structure: 3EUA
- UniProt: O32157
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional references: PubMed
Martin Lehnik-Habrink, Leonie Rempeters, Ákos T Kovács, Christoph Wrede, Claudia Baierlein, Heike Krebber, Oscar P Kuipers, Jörg Stülke
DEAD-Box RNA helicases in Bacillus subtilis have multiple functions and act independently from each other.
J Bacteriol: 2013, 195(3);534-44
[PubMed:23175651]
[WorldCat.org]
[DOI]
(I p)
Alexander K W Elsholz, Kürsad Turgay, Stephan Michalik, Bernd Hessling, Katrin Gronau, Dan Oertel, Ulrike Mäder, Jörg Bernhardt, Dörte Becher, Michael Hecker, Ulf Gerth
Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis.
Proc Natl Acad Sci U S A: 2012, 109(19);7451-6
[PubMed:22517742]
[WorldCat.org]
[DOI]
(I p)
Lehnik-Habrink M, Schaffer M, Mäder U, Diethmaier C, Herzberg C, Stülke J RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y. Mol Microbiol. 2011 81(6): 1459-1473. PubMed:21815947