TasA
- Description: major component of biofilm matrix, forms amyloid fibers
| Gene name | tasA |
| Synonyms | cotN, yqhF |
| Essential | no |
| Product | major component of biofilm matrix |
| Function | biofilm formation |
| Gene expression levels in SubtiExpress: tasA | |
| Interactions involving this protein in SubtInteract: TasA | |
| Regulation of this protein in SubtiPathways: Biofilm, Protein secretion | |
| MW, pI | 28 kDa, 5.442 |
| Gene length, protein length | 783 bp, 261 aa |
| Immediate neighbours | sinR, sipW |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 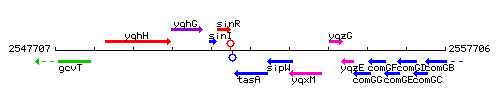
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed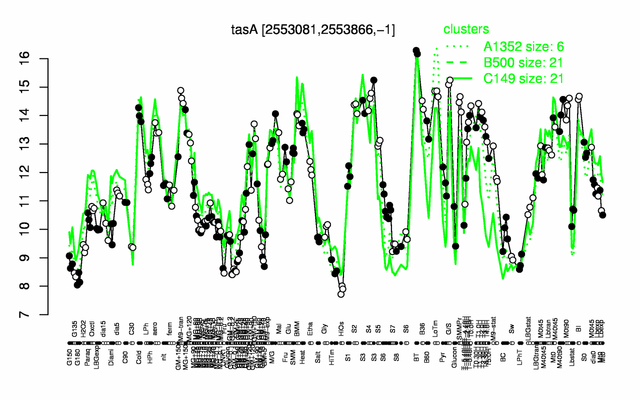
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU24620
Phenotypes of a mutant
- altered cell death pattern in colonies PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: forms amyloid fibers that bind cells together in the biofilm PubMed
- Protein family: peptidase M73 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P54507
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Adam Driks
Tapping into the biofilm: insights into assembly and disassembly of a novel amyloid fibre in Bacillus subtilis.
Mol Microbiol: 2011, 80(5);1133-6
[PubMed:21488983]
[WorldCat.org]
[DOI]
(I p)
Massimiliano Marvasi, Pieter T Visscher, Lilliam Casillas Martinez
Exopolymeric substances (EPS) from Bacillus subtilis: polymers and genes encoding their synthesis.
FEMS Microbiol Lett: 2010, 313(1);1-9
[PubMed:20735481]
[WorldCat.org]
[DOI]
(I p)
Yunrong Chai, Thomas Norman, Roberto Kolter, Richard Losick
An epigenetic switch governing daughter cell separation in Bacillus subtilis.
Genes Dev: 2010, 24(8);754-65
[PubMed:20351052]
[WorldCat.org]
[DOI]
(I p)
Original publications
Diego Romero, Edgardo Sanabria-Valentín, Hera Vlamakis, Roberto Kolter
Biofilm inhibitors that target amyloid proteins.
Chem Biol: 2013, 20(1);102-10
[PubMed:23352144]
[WorldCat.org]
[DOI]
(I p)
Munehiro Asally, Mark Kittisopikul, Pau Rué, Yingjie Du, Zhenxing Hu, Tolga Çağatay, Andra B Robinson, Hongbing Lu, Jordi Garcia-Ojalvo, Gürol M Süel
Localized cell death focuses mechanical forces during 3D patterning in a biofilm.
Proc Natl Acad Sci U S A: 2012, 109(46);18891-6
[PubMed:23012477]
[WorldCat.org]
[DOI]
(I p)
Lehnik-Habrink M, Schaffer M, Mäder U, Diethmaier C, Herzberg C, Stülke J RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y. Mol Microbiol. 2011 81(6): 1459-1473. PubMed:21815947
Diethmaier C, Pietack N, Gunka K, Wrede C, Lehnik-Habrink M, Herzberg C, Hübner S, Stülke J A Novel Factor Controlling Bistability in Bacillus subtilis: The YmdB Protein Affects Flagellin Expression and Biofilm Formation. J Bacteriol.: 2011, 193(21):5997-6007. PubMed:21856853
Diego Romero, Claudio Aguilar, Richard Losick, Roberto Kolter
Amyloid fibers provide structural integrity to Bacillus subtilis biofilms.
Proc Natl Acad Sci U S A: 2010, 107(5);2230-4
[PubMed:20080671]
[WorldCat.org]
[DOI]
(I p)
Birgit Voigt, Haike Antelmann, Dirk Albrecht, Armin Ehrenreich, Karl-Heinz Maurer, Stefan Evers, Gerhard Gottschalk, Jan Maarten van Dijl, Thomas Schweder, Michael Hecker
Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis.
J Mol Microbiol Biotechnol: 2009, 16(1-2);53-68
[PubMed:18957862]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Kazuo Kobayashi
SlrR/SlrA controls the initiation of biofilm formation in Bacillus subtilis.
Mol Microbiol: 2008, 69(6);1399-410
[PubMed:18647168]
[WorldCat.org]
[DOI]
(I p)
Frances Chu, Daniel B Kearns, Anna McLoon, Yunrong Chai, Roberto Kolter, Richard Losick
A novel regulatory protein governing biofilm formation in Bacillus subtilis.
Mol Microbiol: 2008, 68(5);1117-27
[PubMed:18430133]
[WorldCat.org]
[DOI]
(I p)
Yunrong Chai, Frances Chu, Roberto Kolter, Richard Losick
Bistability and biofilm formation in Bacillus subtilis.
Mol Microbiol: 2008, 67(2);254-63
[PubMed:18047568]
[WorldCat.org]
[DOI]
(P p)
Mark A Strauch, Benjamin G Bobay, John Cavanagh, Fude Yao, Angelo Wilson, Yoann Le Breton
Abh and AbrB control of Bacillus subtilis antimicrobial gene expression.
J Bacteriol: 2007, 189(21);7720-32
[PubMed:17720793]
[WorldCat.org]
[DOI]
(P p)
Steven S Branda, Frances Chu, Daniel B Kearns, Richard Losick, Roberto Kolter
A major protein component of the Bacillus subtilis biofilm matrix.
Mol Microbiol: 2006, 59(4);1229-38
[PubMed:16430696]
[WorldCat.org]
[DOI]
(P p)
Frances Chu, Daniel B Kearns, Steven S Branda, Roberto Kolter, Richard Losick
Targets of the master regulator of biofilm formation in Bacillus subtilis.
Mol Microbiol: 2006, 59(4);1216-28
[PubMed:16430695]
[WorldCat.org]
[DOI]
(P p)
Ulrike Mäder, Georg Homuth, Christian Scharf, Knut Büttner, Rüdiger Bode, Michael Hecker
Transcriptome and proteome analysis of Bacillus subtilis gene expression modulated by amino acid availability.
J Bacteriol: 2002, 184(15);4288-95
[PubMed:12107147]
[WorldCat.org]
[DOI]
(P p)
A G Stöver, A Driks
Regulation of synthesis of the Bacillus subtilis transition-phase, spore-associated antibacterial protein TasA.
J Bacteriol: 1999, 181(17);5476-81
[PubMed:10464223]
[WorldCat.org]
[DOI]
(P p)
M Serrano, R Zilhão, E Ricca, A J Ozin, C P Moran, A O Henriques
A Bacillus subtilis secreted protein with a role in endospore coat assembly and function.
J Bacteriol: 1999, 181(12);3632-43
[PubMed:10368135]
[WorldCat.org]
[DOI]
(P p)
A G Stöver, A Driks
Secretion, localization, and antibacterial activity of TasA, a Bacillus subtilis spore-associated protein.
J Bacteriol: 1999, 181(5);1664-72
[PubMed:10049401]
[WorldCat.org]
[DOI]
(P p)