AbrB
Revision as of 12:46, 2 April 2014 by 134.76.38.147 (talk)
- Description: transcriptional regulator of transition state genes
| Gene name | abrB |
| Synonyms | cpsX, tolB |
| Essential | no |
| Product | transcriptional regulator |
| Function | regulation of gene expression during the transition from growth to stationary phase |
| Gene expression levels in SubtiExpress: abrB | |
| Interactions involving this protein in SubtInteract: AbrB | |
| Metabolic function and regulation of this protein in SubtiPathways: abrB | |
| MW, pI | 10 kDa, 6.57 |
| Gene length, protein length | 282 bp, 94 aa |
| Immediate neighbours | yabC, metS |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 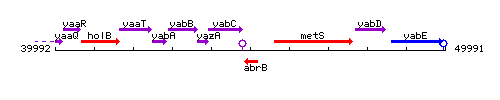
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The AbrB regulon
The gene
Basic information
- Locus tag: BSU00370
Phenotypes of a mutant
No swarming motility on B medium. PubMed
Database entries
- BsubCyc: BSU00370
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
Extended information on the protein
- Kinetic information:
- Modification: phosphorylated on Ser-86 PubMed
Database entries
- BsubCyc: BSU00370
- UniProt: P08874
- KEGG entry: [3]
Additional information
Expression and regulation
- Operon: abrB PubMed
- Regulation: expressed at the onset of stationary phase PubMed
- Additional information:
Biological materials
- Mutant: TT731 (aphA3)
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Richard Losick, Harvard Univ., Cambridge, USA homepage
Mark Strauch, Baltimore, USA homepage
Your additional remarks
References
Reviews
The AbrB regulon:
Other original publications