Difference between revisions of "TasA"
| Line 93: | Line 93: | ||
* '''Regulation:''' repressed by [[SinR]] [http://www.ncbi.nlm.nih.gov/sites/entrez/16430695 PubMed], repressed by casamino acids [http://www.ncbi.nlm.nih.gov/pubmed/12107147 PubMed] | * '''Regulation:''' repressed by [[SinR]] [http://www.ncbi.nlm.nih.gov/sites/entrez/16430695 PubMed], repressed by casamino acids [http://www.ncbi.nlm.nih.gov/pubmed/12107147 PubMed] | ||
| + | ** repressed during logrithmic growth ([[AbrB]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/10464223,17720793 PubMed] | ||
* '''Regulatory mechanism:''' [[SinR]]: transcription repression [http://www.ncbi.nlm.nih.gov/sites/entrez/16430695 PubMed] | * '''Regulatory mechanism:''' [[SinR]]: transcription repression [http://www.ncbi.nlm.nih.gov/sites/entrez/16430695 PubMed] | ||
| + | ** [[AbrB]]: transcription repression [http://www.ncbi.nlm.nih.gov/sites/entrez/10464223,17720793 PubMed] | ||
* '''Additional information:''' | * '''Additional information:''' | ||
Revision as of 11:39, 11 June 2009
- Description: major component of biofilm matrix, translocation-dependent antimicrobial spore component
| Gene name | tasA |
| Synonyms | cotN, yqhF |
| Essential | no |
| Product | major component of biofilm matrix |
| Function | biofilm formation |
| MW, pI | 28 kDa, 5.442 |
| Gene length, protein length | 783 bp, 261 aa |
| Immediate neighbours | sinR, sipW |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 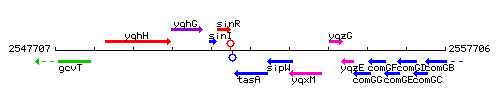
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU24620
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: peptidase M73 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
Database entries
- Structure:
- Swiss prot entry:
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Birgit Voigt, Haike Antelmann, Dirk Albrecht, Armin Ehrenreich, Karl-Heinz Maurer, Stefan Evers, Gerhard Gottschalk, Jan Maarten van Dijl, Thomas Schweder, Michael Hecker
Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis.
J Mol Microbiol Biotechnol: 2009, 16(1-2);53-68
[PubMed:18957862]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Ulrike Mäder, Georg Homuth, Christian Scharf, Knut Büttner, Rüdiger Bode, Michael Hecker
Transcriptome and proteome analysis of Bacillus subtilis gene expression modulated by amino acid availability.
J Bacteriol: 2002, 184(15);4288-95
[PubMed:12107147]
[WorldCat.org]
[DOI]
(P p)
- Voigt et al. (2009) Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis. J Mol Microbiol Biotechnol. 16: 53-68 PubMed
- Mäder et al. (2002) Transcriptome and Proteome Analysis of Bacillus subtilis Gene Expression Modulated by Amino Acid Availability. J. Bacteriol 184: 1844288-4295 PubMed
- Hahne et al. (2008) From complementarity to comprehensiveness - targeting the membrane proteome of growing Bacillus subtilis by divergent approaches. Proteomics 8: 4123-4136 PubMed
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed