Difference between revisions of "FlgE"
(→Expression and regulation) |
|||
| Line 56: | Line 56: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| + | * not essential for pellicle biofilm formation, but mutant is outcompeted by the wild-type strain when competed during pellicle formation {{PubMed|26122431}} | ||
=== Database entries === | === Database entries === | ||
| Line 131: | Line 132: | ||
* '''Mutant:''' | * '''Mutant:''' | ||
| + | ** DS4681 (marker-less in NCIB3610) {{PubMed|22730131}} | ||
| + | ** TB187 ''amyE''::Phy-''sfgfp'' (marker-less in NCIB3610 with constitutive expressed ''sfgfp'') {{PubMed|26122431}} | ||
| + | ** TB203 ''amyE''::Phy-''mKATE2'' (marker-less in NCIB3610 with constitutive expressed ''mKATE2'') {{PubMed|26122431}} | ||
* '''Expression vector:''' | * '''Expression vector:''' | ||
| Line 147: | Line 151: | ||
=References= | =References= | ||
| − | <pubmed>14651647,18957862 18763711, 9657996,8157612,15175317 17850253 24386445 22730131</pubmed> | + | <pubmed> 26122431, 14651647,18957862 18763711, 9657996,8157612,15175317 17850253 24386445 22730131</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 13:10, 2 July 2015
- Description: flagellar hook protein
| Gene name | flgE |
| Synonyms | flgG |
| Essential | no |
| Product | flagellar hook protein |
| Function | movement and chemotaxis |
| Gene expression levels in SubtiExpress: flgE | |
| MW, pI | 27 kDa, 4.734 |
| Gene length, protein length | 792 bp, 264 aa |
| Immediate neighbours | flgD, ylzI |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 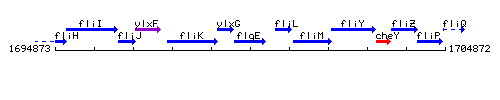
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
motility and chemotaxis, membrane proteins
This gene is a member of the following regulons
CodY regulon, DegU regulon, SigD regulon, Spo0A regulon
The gene
Basic information
- Locus tag: BSU16290
Phenotypes of a mutant
- not essential for pellicle biofilm formation, but mutant is outcompeted by the wild-type strain when competed during pellicle formation PubMed
Database entries
- BsubCyc: BSU16290
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- lack of SigD-dependent gene expression (no hag expression), no motility PubMed
- Protein family: flagella basal body rod proteins family (according to Swiss-Prot)
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: bacterial flagellum basal body (according to Swiss-Prot), extracellular (no signal peptide) PubMed, membrane associated PubMed
Database entries
- BsubCyc: BSU16290
- Structure:
- UniProt: P23446
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: part of the fla-che operon (see there for regulation)
- Regulation: see fla-che operon
- Regulatory mechanism: see fla-che operon
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References