Difference between revisions of "BacD"
| Line 78: | Line 78: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
* '''Modification:''' | * '''Modification:''' | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 109: | Line 109: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=bacD_3870668_3872086_-1 bacD] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=bacD_3870668_3872086_-1 bacD] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' [[SigA]] {{PubMed|19801406}} | + | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|19801406}} |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 139: | Line 139: | ||
=References= | =References= | ||
| − | + | <pubmed>16030213,12372825,19352016,19801406 21948839 22407814 24702628 23317005,12697329,21709425,22298000</pubmed> | |
| − | <pubmed>16030213,12372825,19352016,19801406 21948839 22407814 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 12:39, 8 April 2014
- Description: alanine-anticapsin ligase
| Gene name | bacD |
| Synonyms | ywfE, ipa-83d |
| Essential | no |
| Product | alanine-anticapsin ligase |
| Function | biosynthesis of the antibiotic bacilysin |
| Gene expression levels in SubtiExpress: bacD | |
| MW, pI | 52 kDa, 4.679 |
| Gene length, protein length | 1416 bp, 472 aa |
| Immediate neighbours | bacE, bacC |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 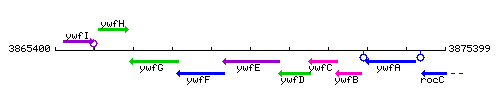
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed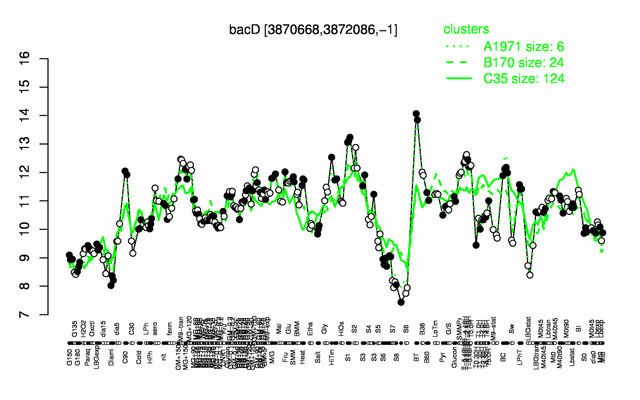
| |
Contents
Categories containing this gene/protein
miscellaneous metabolic pathways, biosynthesis of antibacterial compounds
This gene is a member of the following regulons
AbrB regulon, CodY regulon, ScoC regulon
The gene
Basic information
- Locus tag: BSU37710
Phenotypes of a mutant
Database entries
- BsubCyc: BSU37710
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- ligation of anticapsin to L-Ala to form bacilysin, ultimate step in bacilysin biosynthesis PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- BsubCyc: BSU37710
- UniProt: P39641
- KEGG entry: [2]
- E.C. number: 6.3.2.28
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Takeo Tsuda, Mana Asami, Yoshiaki Koguchi, Shuichi Kojima
Single mutation alters the substrate specificity of L-amino acid ligase.
Biochemistry: 2014, 53(16);2650-60
[PubMed:24702628]
[WorldCat.org]
[DOI]
(I p)
Jared B Parker, Christopher T Walsh
Action and timing of BacC and BacD in the late stages of biosynthesis of the dipeptide antibiotic bacilysin.
Biochemistry: 2013, 52(5);889-901
[PubMed:23317005]
[WorldCat.org]
[DOI]
(I p)
Yasuhito Shomura, Emi Hinokuchi, Hajime Ikeda, Akihiro Senoo, Yuichi Takahashi, Jun-ichi Saito, Hirofumi Komori, Naoki Shibata, Yoshiyuki Yonetani, Yoshiki Higuchi
Structural and enzymatic characterization of BacD, an L-amino acid dipeptide ligase from Bacillus subtilis.
Protein Sci: 2012, 21(5);707-16
[PubMed:22407814]
[WorldCat.org]
[DOI]
(I p)
Takeo Tsuda, Tomomi Suzuki, Shuichi Kojima
Crystallization and preliminary X-ray diffraction analysis of Bacillus subtilis YwfE, an L-amino-acid ligase.
Acta Crystallogr Sect F Struct Biol Cryst Commun: 2012, 68(Pt 2);203-6
[PubMed:22298000]
[WorldCat.org]
[DOI]
(I p)
Takashi Inaoka, Kozo Ochi
Scandium stimulates the production of amylase and bacilysin in Bacillus subtilis.
Appl Environ Microbiol: 2011, 77(22);8181-3
[PubMed:21948839]
[WorldCat.org]
[DOI]
(I p)
Türkan Ebru Köroğlu, Ismail Oğülür, Seval Mutlu, Ayten Yazgan-Karataş, Gülay Ozcengiz
Global regulatory systems operating in Bacilysin biosynthesis in Bacillus subtilis.
J Mol Microbiol Biotechnol: 2011, 20(3);144-55
[PubMed:21709425]
[WorldCat.org]
[DOI]
(I p)
Takashi Inaoka, Guojun Wang, Kozo Ochi
ScoC regulates bacilysin production at the transcription level in Bacillus subtilis.
J Bacteriol: 2009, 191(23);7367-71
[PubMed:19801406]
[WorldCat.org]
[DOI]
(I p)
Kuniki Kino, Yoichi Kotanaka, Toshinobu Arai, Makoto Yagasaki
A novel L-amino acid ligase from Bacillus subtilis NBRC3134, a microorganism producing peptide-antibiotic rhizocticin.
Biosci Biotechnol Biochem: 2009, 73(4);901-7
[PubMed:19352016]
[WorldCat.org]
[DOI]
(I p)
Kazuhiko Tabata, Hajime Ikeda, Shin-Ichi Hashimoto
ywfE in Bacillus subtilis codes for a novel enzyme, L-amino acid ligase.
J Bacteriol: 2005, 187(15);5195-202
[PubMed:16030213]
[WorldCat.org]
[DOI]
(P p)
Ayten Yazgan Karataş, Seçil Cetin, Gülay Ozcengiz
The effects of insertional mutations in comQ, comP, srfA, spo0H, spo0A and abrB genes on bacilysin biosynthesis in Bacillus subtilis.
Biochim Biophys Acta: 2003, 1626(1-3);51-6
[PubMed:12697329]
[WorldCat.org]
(P p)
Takashi Inaoka, Kosaku Takahashi, Mayumi Ohnishi-Kameyama, Mitsuru Yoshida, Kozo Ochi
Guanine nucleotides guanosine 5'-diphosphate 3'-diphosphate and GTP co-operatively regulate the production of an antibiotic bacilysin in Bacillus subtilis.
J Biol Chem: 2003, 278(4);2169-76
[PubMed:12372825]
[WorldCat.org]
[DOI]
(P p)