Difference between revisions of "MutL"
| Line 56: | Line 56: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU17050&redirect=T BSU17050] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/mutSL.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/mutSL.html] | ||
| Line 94: | Line 95: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU17050&redirect=T BSU17050] | ||
* '''Structure:''' [http://www.rcsb.org/pdb/explore/explore.do?pdbId=3KDK 3KDK] (C-terminal domain) {{PubMed|20603082}} | * '''Structure:''' [http://www.rcsb.org/pdb/explore/explore.do?pdbId=3KDK 3KDK] (C-terminal domain) {{PubMed|20603082}} | ||
Revision as of 13:47, 2 April 2014
- Description: DNA mismatch repair
| Gene name | mutL |
| Synonyms | |
| Essential | no |
| Product | DNA mismatch repair |
| Function | DNA repair |
| Gene expression levels in SubtiExpress: mutL | |
| Interactions involving this protein in SubtInteract: MutL | |
| MW, pI | 70 kDa, 5.583 |
| Gene length, protein length | 1881 bp, 627 aa |
| Immediate neighbours | mutS, ymzD |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed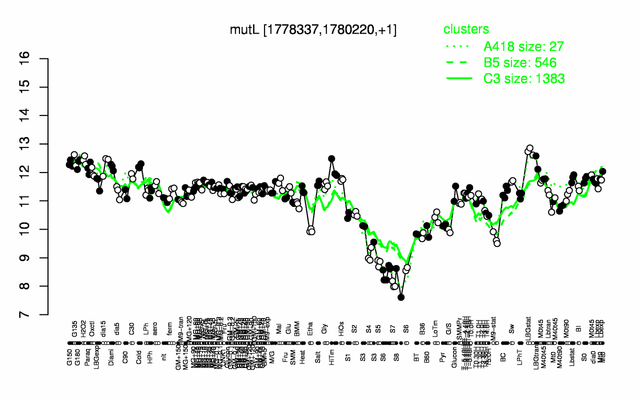
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU17050
Phenotypes of a mutant
Database entries
- BsubCyc: BSU17050
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: DNA mismatch repair mutL/hexB family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- forms foci at midcell position, the frequency of foci increases upon mismatch formation PubMed
Database entries
- BsubCyc: BSU17050
- UniProt: P49850
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Justin S Lenhart, Jeremy W Schroeder, Brian W Walsh, Lyle A Simmons
DNA repair and genome maintenance in Bacillus subtilis.
Microbiol Mol Biol Rev: 2012, 76(3);530-64
[PubMed:22933559]
[WorldCat.org]
[DOI]
(I p)
Original publications
Justin S Lenhart, Monica C Pillon, Alba Guarné, Lyle A Simmons
Trapping and visualizing intermediate steps in the mismatch repair pathway in vivo.
Mol Microbiol: 2013, 90(4);680-98
[PubMed:23998896]
[WorldCat.org]
[DOI]
(I p)
Nina Y Yao, Jeremy W Schroeder, Olga Yurieva, Lyle A Simmons, Mike E O'Donnell
Cost of rNTP/dNTP pool imbalance at the replication fork.
Proc Natl Acad Sci U S A: 2013, 110(32);12942-7
[PubMed:23882084]
[WorldCat.org]
[DOI]
(I p)
Nicholas J Bolz, Justin S Lenhart, Steven C Weindorf, Lyle A Simmons
Residues in the N-terminal domain of MutL required for mismatch repair in Bacillus subtilis.
J Bacteriol: 2012, 194(19);5361-7
[PubMed:22843852]
[WorldCat.org]
[DOI]
(I p)
Katrin Gunka, Stefan Tholen, Jan Gerwig, Christina Herzberg, Jörg Stülke, Fabian M Commichau
A high-frequency mutation in Bacillus subtilis: requirements for the decryptification of the gudB glutamate dehydrogenase gene.
J Bacteriol: 2012, 194(5);1036-44
[PubMed:22178973]
[WorldCat.org]
[DOI]
(I p)
Andrew D Klocko, Jeremy W Schroeder, Brian W Walsh, Justin S Lenhart, Margery L Evans, Lyle A Simmons
Mismatch repair causes the dynamic release of an essential DNA polymerase from the replication fork.
Mol Microbiol: 2011, 82(3);648-63
[PubMed:21958350]
[WorldCat.org]
[DOI]
(I p)
Monica C Pillon, Jeffrey H Miller, Alba Guarné
The endonuclease domain of MutL interacts with the β sliding clamp.
DNA Repair (Amst): 2011, 10(1);87-93
[PubMed:21050827]
[WorldCat.org]
[DOI]
(I p)
Monica C Pillon, Jessica J Lorenowicz, Michael Uckelmann, Andrew D Klocko, Ryan R Mitchell, Yu Seon Chung, Paul Modrich, Graham C Walker, Lyle A Simmons, Peter Friedhoff, Alba Guarné
Structure of the endonuclease domain of MutL: unlicensed to cut.
Mol Cell: 2010, 39(1);145-51
[PubMed:20603082]
[WorldCat.org]
[DOI]
(I p)
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
Mario Pedraza-Reyes, Ronald E Yasbin
Contribution of the mismatch DNA repair system to the generation of stationary-phase-induced mutants of Bacillus subtilis.
J Bacteriol: 2004, 186(19);6485-91
[PubMed:15375129]
[WorldCat.org]
[DOI]
(P p)
F Ginetti, M Perego, A M Albertini, A Galizzi
Bacillus subtilis mutS mutL operon: identification, nucleotide sequence and mutagenesis.
Microbiology (Reading): 1996, 142 ( Pt 8);2021-9
[PubMed:8760914]
[WorldCat.org]
[DOI]
(P p)