Difference between revisions of "CwlS"
| Line 63: | Line 63: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| Line 81: | Line 79: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
| + | ** contains four N-acetylglucosamine-polymer-binding [[LysM domain]]s {{PubMed|24355088,18430080}} | ||
** C-terminal D,L-endopeptidase domain {{PubMed|22139507}} | ** C-terminal D,L-endopeptidase domain {{PubMed|22139507}} | ||
* '''Modification:''' | * '''Modification:''' | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 144: | Line 143: | ||
=References= | =References= | ||
| − | <pubmed>16855244,12850135, 22139507 20817675</pubmed> | + | == Reviews== |
| − | + | <pubmed>18430080</pubmed> | |
| + | == Original publications == | ||
| + | <pubmed>16855244,12850135, 22139507 20817675 24355088</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 14:27, 27 December 2013
- Description: D,L-endopeptidase, peptidoglycan hydrolase for cell separation
| Gene name | cwlS |
| Synonyms | yojL |
| Essential | no |
| Product | D,L-endopeptidase, peptidoglycan hydrolase |
| Function | cell separation |
| Gene expression levels in SubtiExpress: cwlS | |
| MW, pI | 44 kDa, 10.438 |
| Gene length, protein length | 1242 bp, 414 aa |
| Immediate neighbours | yojM, yojK |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 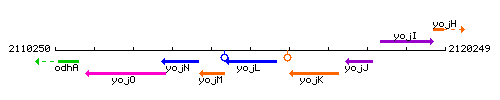
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed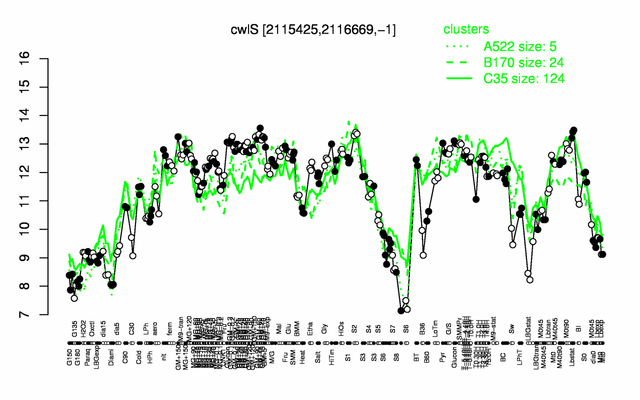
| |
Contents
Categories containing this gene/protein
cell wall degradation/ turnover, membrane proteins
This gene is a member of the following regulons
Abh regulon, AbrB regulon, CcpA regulon, SigD regulon, SigH regulon,
The gene
Basic information
- Locus tag: BSU19410
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Hydrolysis of gamma-D-glutamyl bonds to the L-terminus (position 7) of meso-diaminopimelic acid (meso-A2pm) in 7-(L-Ala-gamma-D-Glu)-meso-A2pm and 7-(L-Ala-gamma-D-Glu)-7-(D-Ala)-meso-A2pm (according to Swiss-Prot)
- Protein family: Cu-Zn superoxide dismutase family (according to Swiss-Prot)
- Paralogous protein(s): the C-terminal D,L-endopeptidase domains of LytE, LytF, CwlS, and CwlO exhibit strong sequence similarity
Extended information on the protein
- Kinetic information:
- Domains:
- contains four N-acetylglucosamine-polymer-binding LysM domains PubMed
- C-terminal D,L-endopeptidase domain PubMed
- Modification:
- Effectors of protein activity:
- Localization:
- cell membrane (according to Swiss-Prot)
- localizes to cell septa and poles PubMed
Database entries
- Structure:
- UniProt: O31852
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor: SigD, SigH, according to PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications