Difference between revisions of "CtaD"
| Line 88: | Line 88: | ||
* '''Modification:''' | * '''Modification:''' | ||
| − | * '''Cofactor(s):''' | + | * '''Cofactor(s):''' Cu(B) {{PubMed|10837475}} |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 146: | Line 146: | ||
=References= | =References= | ||
| − | <pubmed>,1685007,10551842,9829923,17114254 20817675</pubmed> | + | <pubmed>,1685007,10551842,9829923,17114254 20817675 10837475 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 18:24, 11 August 2013
- Description: cytochrome-c oxidase (subunit I)
| Gene name | ctaD |
| Synonyms | |
| Essential | no |
| Product | cytochrome-c oxidase (subunit I) |
| Function | respiration |
| Gene expression levels in SubtiExpress: ctaD | |
| MW, pI | 68 kDa, 6.985 |
| Gene length, protein length | 1866 bp, 622 aa |
| Immediate neighbours | ctaC, ctaE |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 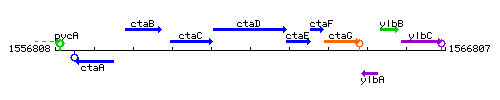
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed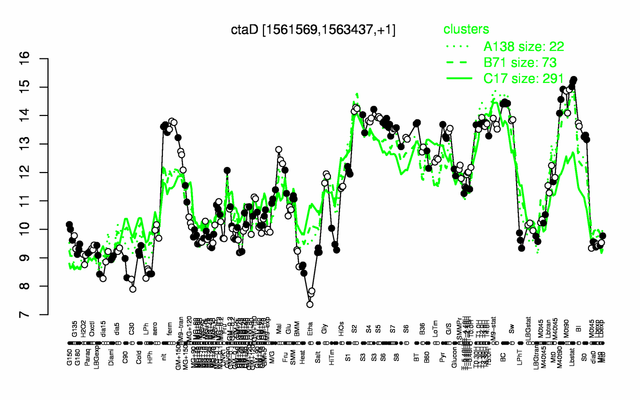
| |
Contents
Categories containing this gene/protein
respiration, membrane proteins
This gene is a member of the following regulons
Abh regulon, AbrB regulon, ResD regulon, Efp-dependent proteins
The gene
Basic information
- Locus tag: BSU14900
Phenotypes of a mutant
essential according to PubMed, non-essential according to PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 4 ferrocytochrome c + O2 + 4 H+ = 4 ferricytochrome c + 2 H2O (according to Swiss-Prot)
- Protein family: heme-copper respiratory oxidase family (according to Swiss-Prot)
- Paralogous protein(s): QoxB
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s): Cu(B) PubMed
- Effectors of protein activity:
- Localization:
- cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P24010
- KEGG entry: [3]
- E.C. number: 1.9.3.1
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
- translation is likely to require Efp due to the presence of several consecutive proline residues PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Onuma Chumsakul, Hiroki Takahashi, Taku Oshima, Takahiro Hishimoto, Shigehiko Kanaya, Naotake Ogasawara, Shu Ishikawa
Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation.
Nucleic Acids Res: 2011, 39(2);414-28
[PubMed:20817675]
[WorldCat.org]
[DOI]
(I p)
Helena B Thomaides, Ella J Davison, Lisa Burston, Hazel Johnson, David R Brown, Alison C Hunt, Jeffery Errington, Lloyd Czaplewski
Essential bacterial functions encoded by gene pairs.
J Bacteriol: 2007, 189(2);591-602
[PubMed:17114254]
[WorldCat.org]
[DOI]
(P p)
N R Mattatall, J Jazairi, B C Hill
Characterization of YpmQ, an accessory protein required for the expression of cytochrome c oxidase in Bacillus subtilis.
J Biol Chem: 2000, 275(37);28802-9
[PubMed:10837475]
[WorldCat.org]
[DOI]
(P p)
N Azarkina, S Siletsky, V Borisov, C von Wachenfeldt, L Hederstedt, A A Konstantinov
A cytochrome bb'-type quinol oxidase in Bacillus subtilis strain 168.
J Biol Chem: 1999, 274(46);32810-7
[PubMed:10551842]
[WorldCat.org]
[DOI]
(P p)
X Liu, H W Taber
Catabolite regulation of the Bacillus subtilis ctaBCDEF gene cluster.
J Bacteriol: 1998, 180(23);6154-63
[PubMed:9829923]
[WorldCat.org]
[DOI]
(P p)
J van der Oost, C von Wachenfeld, L Hederstedt, M Saraste
Bacillus subtilis cytochrome oxidase mutants: biochemical analysis and genetic evidence for two aa3-type oxidases.
Mol Microbiol: 1991, 5(8);2063-72
[PubMed:1685007]
[WorldCat.org]
[DOI]
(P p)