Difference between revisions of "YocH"
| Line 35: | Line 35: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 90: | Line 86: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' | + | * '''[[SubtInteract|Interactions]]:''' |
| − | * '''Localization:''' extracellular (signal peptide) [http://www.ncbi.nlm.nih.gov/pubmed/18957862 PubMed] | + | * '''[[Localization]]:''' |
| + | ** extracellular (signal peptide) [http://www.ncbi.nlm.nih.gov/pubmed/18957862 PubMed] | ||
=== Database entries === | === Database entries === | ||
| Line 112: | Line 109: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=yocH_2092899_2093762_-1 yocH] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=yocH_2092899_2093762_-1 yocH] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' [[SigA]] {{PubMed|12950927}} | + | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|12950927}} |
* '''Regulation:''' | * '''Regulation:''' | ||
Revision as of 21:09, 15 March 2013
- Description: peptidoglycan hydrolase (amidase)
| Gene name | yocH |
| Synonyms | |
| Essential | no |
| Product | peptidoglycan hydrolase |
| Function | cell wall turnover |
| Gene expression levels in SubtiExpress: yocH | |
| MW, pI | 30 kDa, 8.371 |
| Gene length, protein length | 861 bp, 287 aa |
| Immediate neighbours | desR, yocI |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 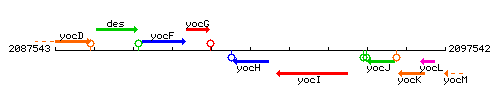
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed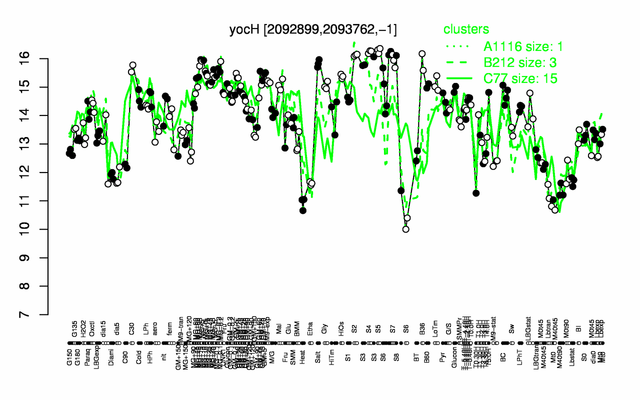
| |
Contents
Categories containing this gene/protein
cell wall degradation/ turnover
This gene is a member of the following regulons
AbrB regulon, Spo0A regulon, WalR regulon
The gene
Basic information
- Locus tag: BSU19210
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- extracellular (signal peptide) PubMed
Database entries
- Structure:
- UniProt: O34669
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: yocH PubMed
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: AH024 knock-out mutant (kan) available in the lab of Kevin Devine
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Ishita M Shah, Jonathan Dworkin
Induction and regulation of a secreted peptidoglycan hydrolase by a membrane Ser/Thr kinase that detects muropeptides.
Mol Microbiol: 2010, 75(5);1232-43
[PubMed:20070526]
[WorldCat.org]
[DOI]
(I p)
Birgit Voigt, Haike Antelmann, Dirk Albrecht, Armin Ehrenreich, Karl-Heinz Maurer, Stefan Evers, Gerhard Gottschalk, Jan Maarten van Dijl, Thomas Schweder, Michael Hecker
Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis.
J Mol Microbiol Biotechnol: 2009, 16(1-2);53-68
[PubMed:18957862]
[WorldCat.org]
[DOI]
(I p)
Mélanie A Hamon, Nicola R Stanley, Robert A Britton, Alan D Grossman, Beth A Lazazzera
Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis.
Mol Microbiol: 2004, 52(3);847-60
[PubMed:15101989]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Masaya Fujita, Shane T Jensen, Patrick Eichenberger, José E González-Pastor, Jun S Liu, Richard Losick
The Spo0A regulon of Bacillus subtilis.
Mol Microbiol: 2003, 50(5);1683-701
[PubMed:14651647]
[WorldCat.org]
[DOI]
(P p)
Alistair Howell, Sarah Dubrac, Kasper Krogh Andersen, David Noone, Juliette Fert, Tarek Msadek, Kevin Devine
Genes controlled by the essential YycG/YycF two-component system of Bacillus subtilis revealed through a novel hybrid regulator approach.
Mol Microbiol: 2003, 49(6);1639-55
[PubMed:12950927]
[WorldCat.org]
[DOI]
(P p)