Difference between revisions of "LiaF"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' inhibitor of [[LiaS]] kinase activity <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || negative effector of [[ | + | |style="background:#ABCDEF;" align="center"| '''Product''' || negative effector of [[LiaS]] |
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || control of [[LiaR]] activity | |style="background:#ABCDEF;" align="center"|'''Function''' || control of [[LiaR]] activity | ||
| Line 35: | Line 35: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 65: | Line 61: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| Line 74: | Line 68: | ||
* '''Catalyzed reaction/ biological activity:''' | * '''Catalyzed reaction/ biological activity:''' | ||
| + | ** inhibitor of [[LiaS]] kinase activity, maintains [[LiaS]] in the phosphatase state in the absence of the stress signal {{PubMed|23279150}} | ||
* '''Protein family:''' | * '''Protein family:''' | ||
| Line 149: | Line 144: | ||
=References= | =References= | ||
'''Additional publications:''' {{PubMed|22092710}} | '''Additional publications:''' {{PubMed|22092710}} | ||
| − | <pubmed>19164152, 20639339,15273097,17660417,16816187,15101989,17660417,16816187 </pubmed> | + | <pubmed>19164152, 20639339,15273097,17660417,16816187,15101989,17660417,16816187 23279150 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 10:31, 3 January 2013
- Description: inhibitor of LiaS kinase activity
| Gene name | liaF |
| Synonyms | yvqF |
| Essential | no |
| Product | negative effector of LiaS |
| Function | control of LiaR activity |
| Gene expression levels in SubtiExpress: liaF | |
| MW, pI | 26 kDa, 9.211 |
| Gene length, protein length | 723 bp, 241 aa |
| Immediate neighbours | liaS, liaG |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 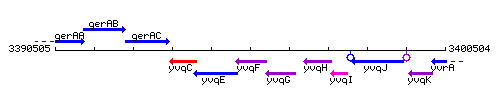
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed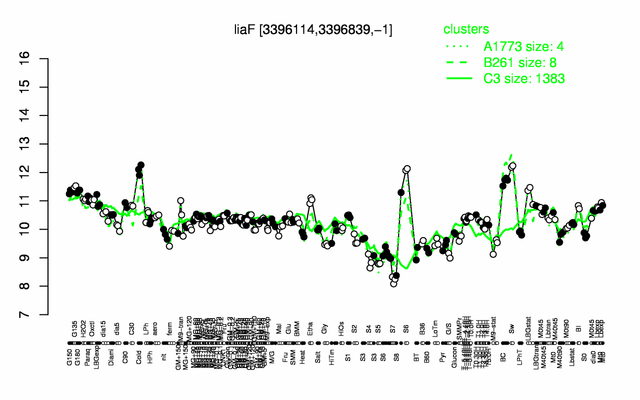
| |
Contents
Categories containing this gene/protein
transcription factors and their control, resistance against oxidative and electrophile stress, resistance against toxins/ antibiotics, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU33100
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: O32199
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Karen Schrecke, Sina Jordan, Thorsten Mascher
Stoichiometry and perturbation studies of the LiaFSR system of Bacillus subtilis.
Mol Microbiol: 2013, 87(4);769-88
[PubMed:23279150]
[WorldCat.org]
[DOI]
(I p)
Diana Wolf, Falk Kalamorz, Tina Wecke, Anna Juszczak, Ulrike Mäder, Georg Homuth, Sina Jordan, Janine Kirstein, Michael Hoppert, Birgit Voigt, Michael Hecker, Thorsten Mascher
In-depth profiling of the LiaR response of Bacillus subtilis.
J Bacteriol: 2010, 192(18);4680-93
[PubMed:20639339]
[WorldCat.org]
[DOI]
(I p)
Anna-Barbara Hachmann, Esther R Angert, John D Helmann
Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin.
Antimicrob Agents Chemother: 2009, 53(4);1598-609
[PubMed:19164152]
[WorldCat.org]
[DOI]
(I p)
Sina Jordan, Eva Rietkötter, Mark A Strauch, Falk Kalamorz, Bronwyn G Butcher, John D Helmann, Thorsten Mascher
LiaRS-dependent gene expression is embedded in transition state regulation in Bacillus subtilis.
Microbiology (Reading): 2007, 153(Pt 8);2530-2540
[PubMed:17660417]
[WorldCat.org]
[DOI]
(P p)
Sina Jordan, Anja Junker, John D Helmann, Thorsten Mascher
Regulation of LiaRS-dependent gene expression in bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system.
J Bacteriol: 2006, 188(14);5153-66
[PubMed:16816187]
[WorldCat.org]
[DOI]
(P p)
Thorsten Mascher, Sara L Zimmer, Terry-Ann Smith, John D Helmann
Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis.
Antimicrob Agents Chemother: 2004, 48(8);2888-96
[PubMed:15273097]
[WorldCat.org]
[DOI]
(P p)
Mélanie A Hamon, Nicola R Stanley, Robert A Britton, Alan D Grossman, Beth A Lazazzera
Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis.
Mol Microbiol: 2004, 52(3);847-60
[PubMed:15101989]
[WorldCat.org]
[DOI]
(P p)