Difference between revisions of "FrlB"
(→Expression and regulation) |
|||
| Line 112: | Line 112: | ||
** ''[[frlB]]-[[frlO]]'' {{PubMed|23175651}} | ** ''[[frlB]]-[[frlO]]'' {{PubMed|23175651}} | ||
** ''[[frlB]]'' {{PubMed|23175651}} | ** ''[[frlB]]'' {{PubMed|23175651}} | ||
| − | |||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=frlB_3351110_3352096_-1 frlB] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=frlB_3351110_3352096_-1 frlB] {{PubMed|22383849}} | ||
Revision as of 17:12, 26 November 2012
- Description: fructoselysine-6-P-glycosidase
| Gene name | frlB |
| Synonyms | yurP |
| Essential | no |
| Product | fructoselysine-6-P-glycosidase |
| Function | metabolism of aminoacylated fructose |
| Gene expression levels in SubtiExpress: frlB | |
| Metabolic function and regulation of this protein in SubtiPathways: Alternative nitrogen sources | |
| MW, pI | 36 kDa, 5.442 |
| Gene length, protein length | 984 bp, 328 aa |
| Immediate neighbours | frlO, yurQ |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 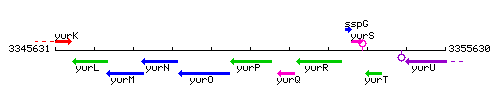
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed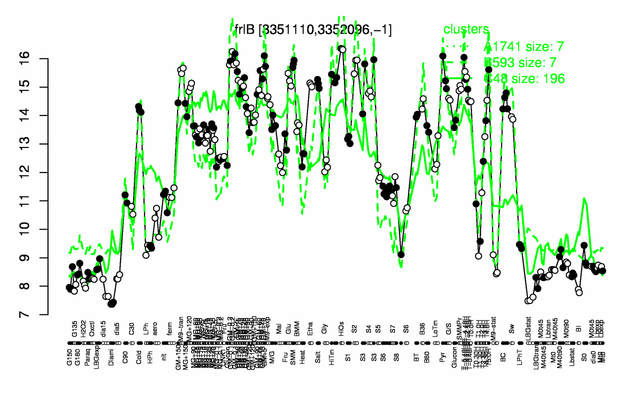
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources, utilization of nitrogen sources other than amino acids, membrane proteins, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU32610
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- phosphorylated on Arg-48 PubMed
- Cofactor(s):
- Effectors of protein activity:
- Localization: membrane associated PubMed
Database entries
- Structure: 3EUA
- UniProt: O32157
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional references: PubMed
Lehnik-Habrink M, Schaffer M, Mäder U, Diethmaier C, Herzberg C, Stülke J RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y. Mol Microbiol. 2011 81(6): 1459-1473. PubMed:21815947
Veronika Maria Deppe, Stephanie Klatte, Johannes Bongaerts, Karl-Heinz Maurer, Timothy O'Connell, Friedhelm Meinhardt
Genetic control of amadori product degradation in Bacillus subtilis via regulation of frlBONMD expression by FrlR.
Appl Environ Microbiol: 2011, 77(9);2839-46
[PubMed:21398478]
[WorldCat.org]
[DOI]
(I p)
Veronika Maria Deppe, Johannes Bongaerts, Timothy O'Connell, Karl-Heinz Maurer, Friedhelm Meinhardt
Enzymatic deglycation of Amadori products in bacteria: mechanisms, occurrence and physiological functions.
Appl Microbiol Biotechnol: 2011, 90(2);399-406
[PubMed:21347729]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Boris R Belitsky, Abraham L Sonenshein
Genetic and biochemical analysis of CodY-binding sites in Bacillus subtilis.
J Bacteriol: 2008, 190(4);1224-36
[PubMed:18083814]
[WorldCat.org]
[DOI]
(I p)
Elsa Wiame, Armelle Duquenne, Ghislain Delpierre, Emile Van Schaftingen
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
FEBS Lett: 2004, 577(3);469-72
[PubMed:15556630]
[WorldCat.org]
[DOI]
(P p)
Ken-ichi Yoshida, Hirotake Yamaguchi, Masaki Kinehara, Yo-hei Ohki, Yoshiko Nakaura, Yasutaro Fujita
Identification of additional TnrA-regulated genes of Bacillus subtilis associated with a TnrA box.
Mol Microbiol: 2003, 49(1);157-65
[PubMed:12823818]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Yoshiko Nakaura, Robert P Shivers, Hirotake Yamaguchi, Richard Losick, Yasutaro Fujita, Abraham L Sonenshein
Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis.
J Bacteriol: 2003, 185(6);1911-22
[PubMed:12618455]
[WorldCat.org]
[DOI]
(P p)