Difference between revisions of "Sandbox"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' large subunit of glutamate synthase <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''gltA'' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' '' | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' '' | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | + | |style="background:#ABCDEF;" align="center"| '''Essential''' || no |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || | + | |style="background:#ABCDEF;" align="center"| '''Product''' || glutamate synthase (large subunit) |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || | + | |style="background:#ABCDEF;" align="center"|'''Function''' || glutamate biosynthesis |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || | + | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 168 kDa, 5.47 |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || | + | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 4560 bp, 1520 amino acids |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[gltC]]'', ''[[gltB]]'' |
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS: | + | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS:CAB13728]+-newId sequences] <br/> (Barbe ''et al.'', 2009)''' |
|- | |- | ||
| − | |colspan="2" | '''Genetic context''' <br/> [[Image: | + | |- |
| + | |- | ||
| + | |colspan="2" | '''Genetic context''' <br/> [[Image:gltA_context.gif]] | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| Line 30: | Line 32: | ||
<br/><br/> | <br/><br/> | ||
| + | |||
=The gene= | =The gene= | ||
| Line 35: | Line 38: | ||
=== Basic information === | === Basic information === | ||
| − | * '''Locus tag:''' | + | * '''Locus tag:''' BSU18450 |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| + | |||
| + | auxotrophic for glutamate | ||
=== Database entries === | === Database entries === | ||
| − | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/ | + | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/gltAB.html] |
| − | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+ | + | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG10811] |
=== Additional information=== | === Additional information=== | ||
| Line 52: | Line 57: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' | + | * '''Catalyzed reaction/ biological activity:''' 2 L-glutamate + NADP<sup>+</sup> = L-glutamine + 2-oxoglutarate + NADPH (according to Swiss-Prot) 2 L-glutamate + NADP(+) <=> L-glutamine + 2-oxoglutarate + NADPH |
| − | * '''Protein family:''' | + | * '''Protein family:''' glutamate synthase family (according to Swiss-Prot) glutamate synthase family |
| − | * '''Paralogous protein(s):''' | + | * '''Paralogous protein(s):''' [[YerD]] |
=== Extended information on the protein === | === Extended information on the protein === | ||
| Line 63: | Line 68: | ||
* '''Domains:''' | * '''Domains:''' | ||
| + | ** Glutamine amidotransferase type-2 domain (22-415) | ||
| + | ** Nucleotide binding domain (1060-1112) | ||
* '''Modification:''' | * '''Modification:''' | ||
| − | * '''Cofactor(s):''' | + | * '''Cofactor(s):''' 3Fe-4S, FAD, FMN |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 72: | Line 79: | ||
* '''Interactions:''' | * '''Interactions:''' | ||
| − | * '''Localization:''' | + | * '''Localization:''' membrane associated [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+18763711 PubMed], cytoplasm |
=== Database entries === | === Database entries === | ||
| Line 78: | Line 85: | ||
* '''Structure:''' | * '''Structure:''' | ||
| − | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/ | + | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P39812 P39812] |
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+ | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU18450] |
| − | * '''E.C. number:''' | + | * '''E.C. number:''' [http://www.expasy.org/enzyme/1.4.1.13 1.4.1.13] 3 1.4.1.13] |
=== Additional information=== | === Additional information=== | ||
| + | |||
| + | subject to Clp-dependent proteolysis upon glucose starvation [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+17981983 PubMed] | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' | + | * '''Operon:''' ''[[gltA]]-[[gltB]]'' |
| − | * '''[[Sigma factor]]:''' | + | * '''[[Sigma factor]]:''' [[SigA]] |
| − | * '''Regulation:''' | + | * '''Regulation:''' expression activated by glucose (11 fold) [http://www.ncbi.nlm.nih.gov/pubmed/12850135 PubMed], induced by sugar [http://www.ncbi.nlm.nih.gov/sites/entrez/14523131 PubMed], repressed by arginine [http://www.ncbi.nlm.nih.gov/sites/entrez/15150225 PubMed], ammonium required [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+11029411 PubMed] |
| − | * '''Regulatory mechanism:''' | + | * '''Regulatory mechanism:''' Activator: [[GltC]] [http://www.ncbi.nlm.nih.gov/sites/entrez/2548995 PubMed]; Repressor: [[TnrA]] [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+11029411 PubMed], pos. regulated by a mutant form of [[GltR]] [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+9023181 PubMed] |
| − | * '''Additional information:''' | + | * '''Additional information:''' subject to Clp-dependent proteolysis upon glucose starvation [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+17981983 PubMed] |
=Biological materials = | =Biological materials = | ||
| − | * '''Mutant:''' | + | * '''Mutant:''' GP807 (del ''gltAB''::''tet''), GP222 (''gltA'' under the control of p-xyl), available in [[Stülke]] lab |
* '''Expression vector:''' | * '''Expression vector:''' | ||
| − | + | ||
| − | * '''lacZ fusion:''' | + | * '''lacZ fusion:''' pGP526 (in [[pAC7]]), pGP919 (in [[pAC5]]), available in [[Stülke]] lab |
* '''GFP fusion:''' | * '''GFP fusion:''' | ||
| − | |||
| − | |||
* '''Antibody:''' | * '''Antibody:''' | ||
| Line 114: | Line 121: | ||
=Labs working on this gene/protein= | =Labs working on this gene/protein= | ||
| − | [[ | + | |
| + | |||
| + | |||
| + | [[Linc Sonenshein|Linc Sonenshein]], Tufts University, Boston, MA, USA [http://www.tufts.edu/sackler/microbiology/faculty/sonenshein/index.html Homepage] | ||
| + | |||
| + | [[Stülke|Jörg Stülke]], University of Göttingen, Germany | ||
| + | [http://wwwuser.gwdg.de/~genmibio/stuelke.html Homepage] | ||
=Your additional remarks= | =Your additional remarks= | ||
| Line 120: | Line 133: | ||
=References= | =References= | ||
| − | <pubmed> | + | <pubmed>12850135 7559360 15150225 2548995 17183217 17608797 17134717 14523131, </pubmed> |
| − | # | + | # Blencke et al. (2003) Transcriptional profiling of gene expression in response to glucose in ''Bacillus subtilis'': regulation of the central metabolic pathways. ''Metab Eng.'' '''5:''' 133-149 [http://www.ncbi.nlm.nih.gov/pubmed/12850135 PubMed] |
| − | # | + | # Yoshida K, et al. (2003) Identification of additional TnrA-regulated genes of Bacillus subtilis associated with a TnrA box. ''Mol Microbiol'' '''49(1):''' 157-65. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+12823818 PubMed] |
| − | # | + | # Belitsky, B. R., and Sonenshein, A. L. (1995) Mutations in GltC that increase ''Bacillus subtilis gltA'' expression. J Bacteriol 177: 5696-5700.[http://www.ncbi.nlm.nih.gov/sites/entrez/7559360 PubMed] |
| − | # | + | # Belitsky, B. R., and Sonenshein, A. L. (2004) Modulation of activity of ''Bacillus subtilis'' regulatory proteins GltC and TnrA by glutamate dehydrogenase. J Bacteriol 186: 3399-3407.[http://www.ncbi.nlm.nih.gov/sites/entrez/15150225 PubMed] |
| − | # | + | # Bohannon, D. E., and Sonenshein, A. L. (1989) Positive regulation of glutamate biosynthesis in ''Bacillus subtilis''. J Bacteriol 171: 4718-4727.[http://www.ncbi.nlm.nih.gov/sites/entrez/2548995 PubMed] |
| − | # | + | # Commichau, F. M., Wacker, I., Schleider, J., Blencke, H.-M., Reif, I., Tripal, P., and Stülke, J. (2007) Characterization of ''Bacillus subtilis'' mutants with carbon source-independent glutamate biosynthesis. J Mol Microbiol Biotechnol 12: 106-113. [http://www.ncbi.nlm.nih.gov/sites/entrez/17183217 PubMed] |
| − | # | + | # Commichau, F. M., Herzberg, C., Tripal, P., Valerius, O., and Stülke, J. (2007) A regulatory protein-protein interaction governs glutamate biosynthesis in ''Bacillus subtilis'': The glutamate dehydrogenase RocG moonlights in controlling the transcription factor GltC. Mol Microbiol 65: 642-654. [http://www.ncbi.nlm.nih.gov/sites/entrez/17608797 PubMed] |
| − | # | + | # Picossi, S., Belitsky, B. R., and Sonenshein, A. L. (2007) Molecular mechanism of the regulation of ''Bacillus subtilis gltAB'' expression by GltC. J Mol Biol 365: 1298-1313. [http://www.ncbi.nlm.nih.gov/sites/entrez/17134717 PubMed] |
| − | # | + | # Wacker, I., Ludwig, H., Reif, I., Blencke, H. M., Detsch, C., and Stülke, J. (2003) The regulatory link between carbon and nitrogen metabolism in ''Bacillus subtilis'': regulation of the ''gltAB'' operon by the catabolite control protein CcpA. Microbiology 149: 3001-3009.[http://www.ncbi.nlm.nih.gov/sites/entrez/14523131 PubMed] |
| + | # Belitsky BR, Sonenshein AL (1997) Altered transcription activation specificity of a mutant form of Bacillus subtilis GltR, a LysR family member. J Bacteriol 179:1035-1043 [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+11029411 PubMed] | ||
| + | # Belitsky, B. R., Wray, L. V., Jr., Fisher, S. H., Bohannon, D. E. & Sonenshein, A. L. (2000). Role of TnrA in nitrogen source-dependent repression of Bacillus subtilis glutamate synthase gene expression. J Bacteriol 182, 5939-5947. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+11029411 PubMed] | ||
| + | # Commichau, F. M., Gunka, K., Landmann, J. J. & Stülke, J. (2008) Glutamate metabolism in Bacillus subtilis: Gene expression and enzyme activities evolved to avoid futile cycles and to allow rapid responses to perturbations in the system. J. Bacteriol. 190: 3557-3564. [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+18326565 PubMed] | ||
| + | # Gerth et al. (2008) Clp-dependent proteolysis down-regulates central metabolic pathways in glucose-starved Bacillus subtilis. J Bacteriol 190:321-331 [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+17981983 PubMed] | ||
| + | # Hahne et al. (2008) From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches. Proteomics 8:4123-4136 [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=+18763711 PubMed] | ||
Revision as of 14:04, 8 June 2009
- Description: large subunit of glutamate synthase
| Gene name | gltA |
| Synonyms | |
| Essential | no |
| Product | glutamate synthase (large subunit) |
| Function | glutamate biosynthesis |
| MW, pI | 168 kDa, 5.47 |
| Gene length, protein length | 4560 bp, 1520 amino acids |
| Immediate neighbours | gltC, gltB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 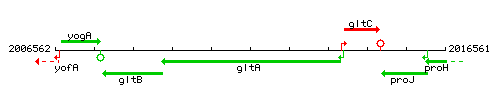
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU18450
Phenotypes of a mutant
auxotrophic for glutamate
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 2 L-glutamate + NADP+ = L-glutamine + 2-oxoglutarate + NADPH (according to Swiss-Prot) 2 L-glutamate + NADP(+) <=> L-glutamine + 2-oxoglutarate + NADPH
- Protein family: glutamate synthase family (according to Swiss-Prot) glutamate synthase family
- Paralogous protein(s): YerD
Extended information on the protein
- Kinetic information:
- Domains:
- Glutamine amidotransferase type-2 domain (22-415)
- Nucleotide binding domain (1060-1112)
- Modification:
- Cofactor(s): 3Fe-4S, FAD, FMN
- Effectors of protein activity:
- Interactions:
- Localization: membrane associated PubMed, cytoplasm
Database entries
- Structure:
- Swiss prot entry: P39812
- KEGG entry: [3]
- E.C. number: 1.4.1.13 3 1.4.1.13]
Additional information
subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Regulation: expression activated by glucose (11 fold) PubMed, induced by sugar PubMed, repressed by arginine PubMed, ammonium required PubMed
- Regulatory mechanism: Activator: GltC PubMed; Repressor: TnrA PubMed, pos. regulated by a mutant form of GltR PubMed
- Additional information: subject to Clp-dependent proteolysis upon glucose starvation PubMed
Biological materials
- Mutant: GP807 (del gltAB::tet), GP222 (gltA under the control of p-xyl), available in Stülke lab
- Expression vector:
- GFP fusion:
- Antibody:
Labs working on this gene/protein
Linc Sonenshein, Tufts University, Boston, MA, USA Homepage
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References
- Blencke et al. (2003) Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways. Metab Eng. 5: 133-149 PubMed
- Yoshida K, et al. (2003) Identification of additional TnrA-regulated genes of Bacillus subtilis associated with a TnrA box. Mol Microbiol 49(1): 157-65. PubMed
- Belitsky, B. R., and Sonenshein, A. L. (1995) Mutations in GltC that increase Bacillus subtilis gltA expression. J Bacteriol 177: 5696-5700.PubMed
- Belitsky, B. R., and Sonenshein, A. L. (2004) Modulation of activity of Bacillus subtilis regulatory proteins GltC and TnrA by glutamate dehydrogenase. J Bacteriol 186: 3399-3407.PubMed
- Bohannon, D. E., and Sonenshein, A. L. (1989) Positive regulation of glutamate biosynthesis in Bacillus subtilis. J Bacteriol 171: 4718-4727.PubMed
- Commichau, F. M., Wacker, I., Schleider, J., Blencke, H.-M., Reif, I., Tripal, P., and Stülke, J. (2007) Characterization of Bacillus subtilis mutants with carbon source-independent glutamate biosynthesis. J Mol Microbiol Biotechnol 12: 106-113. PubMed
- Commichau, F. M., Herzberg, C., Tripal, P., Valerius, O., and Stülke, J. (2007) A regulatory protein-protein interaction governs glutamate biosynthesis in Bacillus subtilis: The glutamate dehydrogenase RocG moonlights in controlling the transcription factor GltC. Mol Microbiol 65: 642-654. PubMed
- Picossi, S., Belitsky, B. R., and Sonenshein, A. L. (2007) Molecular mechanism of the regulation of Bacillus subtilis gltAB expression by GltC. J Mol Biol 365: 1298-1313. PubMed
- Wacker, I., Ludwig, H., Reif, I., Blencke, H. M., Detsch, C., and Stülke, J. (2003) The regulatory link between carbon and nitrogen metabolism in Bacillus subtilis: regulation of the gltAB operon by the catabolite control protein CcpA. Microbiology 149: 3001-3009.PubMed
- Belitsky BR, Sonenshein AL (1997) Altered transcription activation specificity of a mutant form of Bacillus subtilis GltR, a LysR family member. J Bacteriol 179:1035-1043 PubMed
- Belitsky, B. R., Wray, L. V., Jr., Fisher, S. H., Bohannon, D. E. & Sonenshein, A. L. (2000). Role of TnrA in nitrogen source-dependent repression of Bacillus subtilis glutamate synthase gene expression. J Bacteriol 182, 5939-5947. PubMed
- Commichau, F. M., Gunka, K., Landmann, J. J. & Stülke, J. (2008) Glutamate metabolism in Bacillus subtilis: Gene expression and enzyme activities evolved to avoid futile cycles and to allow rapid responses to perturbations in the system. J. Bacteriol. 190: 3557-3564. PubMed
- Gerth et al. (2008) Clp-dependent proteolysis down-regulates central metabolic pathways in glucose-starved Bacillus subtilis. J Bacteriol 190:321-331 PubMed
- Hahne et al. (2008) From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches. Proteomics 8:4123-4136 PubMed