Difference between revisions of "FruA"
| Line 72: | Line 72: | ||
* '''Interactions:''' | * '''Interactions:''' | ||
| − | * '''Localization:''' Membrane (Homogeneous) [http://www.ncbi.nlm.nih.gov/sites/entrez/16479537 PubMed] | + | * '''Localization:''' cell membrane (according to Swiss-Prot), Membrane (Homogeneous) [http://www.ncbi.nlm.nih.gov/sites/entrez/16479537 PubMed] |
=== Database entries === | === Database entries === | ||
Revision as of 15:28, 30 April 2009
- Description: fructose-specific phosphotransferase system, EIIABC
| Gene name | fruA |
| Synonyms | |
| Essential | no |
| Product | fructose-specific phosphotransferase system, EIIABC |
| Function | fructose uptake and phosphorylation |
| MW, pI | 67 kDa, 5.241 |
| Gene length, protein length | 1905 bp, 635 aa |
| Immediate neighbours | fruK, sipT |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 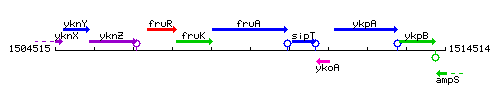
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: PTS permease, fructose/ mannitol permease (Fru) family PubMed
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cell membrane (according to Swiss-Prot), Membrane (Homogeneous) PubMed
Database entries
- Structure:
- Swiss prot entry:
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
- Reizer et al. (1999) Novel phosphotransferase system genes revealed by genome analysis - the complete complement of PTS proteins encoded within the genome of Bacillus subtilis. Microbiology 145: 3419-3429 PubMed
- Meile et al. (2006) Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory Proteomics 6: 2135-2146. PubMed
- Reizer, J., Bachem, S., Reizer, A., Arnaud, M., Saier Jr., M. H. & Stülke, J. (1999) Novel phosphotransferase system genes revealed by genome analysis – the complete complement of PTS proteins encoded within the genome of Bacillus subtilis. Microbiology 145: 3419-3429. PubMed
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed