Difference between revisions of "GltC"
(→Phenotypes of a mutant) |
|||
| Line 33: | Line 33: | ||
=The gene= | =The gene= | ||
| − | == Basic information == | + | === Basic information === |
* '''Coordinates:''' | * '''Coordinates:''' | ||
| − | ==Phenotypes of a mutant == | + | ===Phenotypes of a mutant === |
''gltC'' mutants are auxotrophic for glutamate. | ''gltC'' mutants are auxotrophic for glutamate. | ||
| − | == Database entries == | + | === Database entries === |
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/gltC.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/gltC.html] | ||
| Line 46: | Line 46: | ||
* '''SubtiList entry:'''[http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG10810] | * '''SubtiList entry:'''[http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG10810] | ||
| − | == Additional information== | + | === Additional information=== |
=The protein= | =The protein= | ||
| − | == Basic information/ Evolution == | + | === Basic information/ Evolution === |
* '''Catalyzed reaction/ biological activity:''' transcription activation of the ''gltAB'' operon [http://www.ncbi.nlm.nih.gov/sites/entrez/2548995 PubMed] | * '''Catalyzed reaction/ biological activity:''' transcription activation of the ''gltAB'' operon [http://www.ncbi.nlm.nih.gov/sites/entrez/2548995 PubMed] | ||
| Line 58: | Line 58: | ||
* '''Paralogous protein(s):''' none, but there are 19 members of the LysR family in ''B. subtilis'' | * '''Paralogous protein(s):''' none, but there are 19 members of the LysR family in ''B. subtilis'' | ||
| − | == Extended information on the protein == | + | === Extended information on the protein === |
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| Line 75: | Line 75: | ||
* '''Localization:''' | * '''Localization:''' | ||
| − | == Database entries == | + | === Database entries === |
* '''Structure:''' | * '''Structure:''' | ||
| Line 83: | Line 83: | ||
* '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU18460 KEGG] | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU18460 KEGG] | ||
| − | == Additional information== | + | === Additional information=== |
=Expression and regulation= | =Expression and regulation= | ||
Revision as of 13:58, 17 December 2008
- Description: Transcriptional activator of the gltAB operon. Activates expression of the operon in the absence of arginine.
| Gene name | gltC |
| Synonyms | |
| Essential | No |
| Product | transcriptional regulator (LysR family) |
| Function | positive regulation of the glutamate synthase operon (gltAB) |
| pI, MW | 5.618, 33866.6 Da |
| Gene length, protein length | 900 bp, 300 amino acids |
| Immediate neighbours | gltA, proJ |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 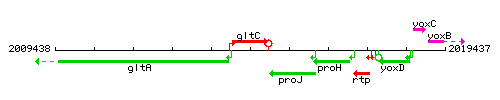
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
gltC mutants are auxotrophic for glutamate.
Database entries
- DBTBS entry: [1]
- SubtiList entry:[2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: transcription activation of the gltAB operon PubMed
- Protein family: LysR-type transcription regulator PubMed
- Paralogous protein(s): none, but there are 19 members of the LysR family in B. subtilis
Extended information on the protein
- Kinetic information:
- Domains: DNA-binding helix-turn-helix motif: AA 18 ... 37
- Modification:
- Cofactor(s):
- Effectors of protein activity: 2-oxoglutarate stimulates transcription activation, glutamate inhibits transcription activation PubMed
- Interactions:
- Localization:
Database entries
- Structure:
- Swiss prot entry: P20668
- KEGG entry: KEGG
Additional information
Expression and regulation
- Operon: gltC
- Sigma factor: SigA
- Regulation: autoregulation by GltC
- Regulatory mechanism: autorepression
- Database entries: DBTBS
- Additional information:
Biological materials
- Mutant: GP344 (erm), GP738 (spc) (available in Stülke lab)
- Expression vector: pGP903 (N-terminal His-tag) (available in Stülke lab)
- lacZ fusion:
- GFP fusion:
- Antibody: available in Stülke lab
Labs working on this gene/protein
Linc Sonenshein, Tufts University, Boston, MA, USA Homepage
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References
- Belitsky, B. R., and Sonenshein, A. L. (1995) Mutations in GltC that increase Bacillus subtilis gltA expression. J Bacteriol 177: 5696-5700.PubMed
- Belitsky, B. R., and Sonenshein, A. L. (2004) Modulation of activity of Bacillus subtilis regulatory proteins GltC and TnrA by glutamate dehydrogenase. J Bacteriol 186: 3399-3407.PubMed
- Bohannon, D. E., and Sonenshein, A. L. (1989) Positive regulation of glutamate biosynthesis in Bacillus subtilis. J Bacteriol 171: 4718-4727.PubMed
- Commichau, F. M., Wacker, I., Schleider, J., Blencke, H.-M., Reif, I., Tripal, P., and Stülke, J. (2007) Characterization of Bacillus subtilis mutants with carbon source-independent glutamate biosynthesis. J Mol Microbiol Biotechnol 12: 106-113. PubMed
- Commichau, F. M., Herzberg, C., Tripal, P., Valerius, O., and Stülke, J. (2007) A regulatory protein-protein interaction governs glutamate biosynthesis in Bacillus subtilis: The glutamate dehydrogenase RocG moonlights in controlling the transcription factor GltC. Mol Microbiol 65: 642-654. PubMed
- Picossi, S., Belitsky, B. R., and Sonenshein, A. L. (2007) Molecular mechanism of the regulation of Bacillus subtilis gltAB expression by GltC. J Mol Biol 365: 1298-1313. PubMed
- Wacker, I., Ludwig, H., Reif, I., Blencke, H. M., Detsch, C., and Stülke, J. (2003) The regulatory link between carbon and nitrogen metabolism in Bacillus subtilis: regulation of the gltAB operon by the catabolite control protein CcpA. Microbiology 149: 3001-3009.PubMed