Difference between revisions of "YtsJ"
| Line 119: | Line 119: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
** belongs to the 100 [[most abundant proteins]] {{PubMed|15378759}} | ** belongs to the 100 [[most abundant proteins]] {{PubMed|15378759}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 3654 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (complex medium with amino acids, without glucose): 4845 {{PubMed|24696501}} | ||
=Biological materials = | =Biological materials = | ||
Revision as of 09:29, 17 April 2014
- Description: malic enzyme, forms a transhydrogenation cycle with MalS for balancing of NADPH
| Gene name | ytsJ |
| Synonyms | |
| Essential | no |
| Product | NADP-dependent malate dehydrogenase |
| Function | malate utilization |
| Gene expression levels in SubtiExpress: ytsJ | |
| Metabolic function and regulation of this protein in SubtiPathways: ytsJ | |
| MW, pI | 43 kDa, 5.046 |
| Gene length, protein length | 1230 bp, 410 aa |
| Immediate neighbours | accD, dnaE |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 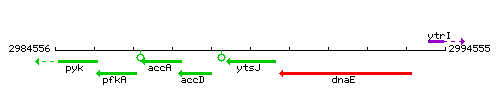
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources, most abundant proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU29220
Phenotypes of a mutant
Poor growth with malate as single carbon source PubMed
Database entries
- BsubCyc: BSU29220
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: (S)-malate + NADP+ = pyruvate + CO2 + NADPH (according to Swiss-Prot) malate <--> pyruvate
- Protein family: malic enzymes family (according to Swiss-Prot)
- Paralogous protein(s): MleA
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Localization:
- Cytoplasm (Homogeneous) PubMed
Database entries
- BsubCyc: BSU29220
- Structure: 1WW8 (from Pyrococcus horikoshii, 46% identity, 63% similarity)
- UniProt: O34962
- KEGG entry: [3]
- E.C. number: 1.1.1.38
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
- belongs to the 100 most abundant proteins PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 3654 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 4845 PubMed
Biological materials
- Mutant:
- GP612 (spc), available in Jörg Stülke's lab
- GP1143 (spc), available in Jörg Stülke's lab
- Expression vector:
- lacZ fusion:
- two-hybrid system:
- FLAG-tag construct: GP1131 (spc, based on pGP1331), available in Jörg Stülke's lab
- Antibody:
Labs working on this gene/protein
Stephane Aymerich, Microbiology and Molecular Genetics, INRA Paris-Grignon, France
Your additional remarks
References
Frederik M Meyer, Jörg Stülke
Malate metabolism in Bacillus subtilis: distinct roles for three classes of malate-oxidizing enzymes.
FEMS Microbiol Lett: 2013, 339(1);17-22
[PubMed:23136871]
[WorldCat.org]
[DOI]
(I p)
Martin Rühl, Dominique Le Coq, Stéphane Aymerich, Uwe Sauer
13C-flux analysis reveals NADPH-balancing transhydrogenation cycles in stationary phase of nitrogen-starving Bacillus subtilis.
J Biol Chem: 2012, 287(33);27959-70
[PubMed:22740702]
[WorldCat.org]
[DOI]
(I p)
Guillaume Lerondel, Thierry Doan, Nicola Zamboni, Uwe Sauer, Stéphane Aymerich
YtsJ has the major physiological role of the four paralogous malic enzyme isoforms in Bacillus subtilis.
J Bacteriol: 2006, 188(13);4727-36
[PubMed:16788182]
[WorldCat.org]
[DOI]
(P p)
Jean-Christophe Meile, Ling Juan Wu, S Dusko Ehrlich, Jeff Errington, Philippe Noirot
Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory.
Proteomics: 2006, 6(7);2135-46
[PubMed:16479537]
[WorldCat.org]
[DOI]
(P p)
Christine Eymann, Annette Dreisbach, Dirk Albrecht, Jörg Bernhardt, Dörte Becher, Sandy Gentner, Le Thi Tam, Knut Büttner, Gerrit Buurman, Christian Scharf, Simone Venz, Uwe Völker, Michael Hecker
A comprehensive proteome map of growing Bacillus subtilis cells.
Proteomics: 2004, 4(10);2849-76
[PubMed:15378759]
[WorldCat.org]
[DOI]
(P p)
Thierry Doan, Pascale Servant, Shigeo Tojo, Hirotake Yamaguchi, Guillaume Lerondel, Ken-Ichi Yoshida, Yasutaro Fujita, Stéphane Aymerich
The Bacillus subtilis ywkA gene encodes a malic enzyme and its transcription is activated by the YufL/YufM two-component system in response to malate.
Microbiology (Reading): 2003, 149(Pt 9);2331-2343
[PubMed:12949160]
[WorldCat.org]
[DOI]
(P p)
Alia Lapidus, Nathalie Galleron, Alexei Sorokin, S Dusko Ehrlich
Sequencing and functional annotation of the Bacillus subtilis genes in the 200 kb rrnB-dnaB region.
Microbiology (Reading): 1997, 143 ( Pt 11);3431-3441
[PubMed:9387221]
[WorldCat.org]
[DOI]
(P p)