Difference between revisions of "SigW"
(→The SigW regulon) |
|||
| Line 150: | Line 150: | ||
<pubmed></pubmed> | <pubmed></pubmed> | ||
==The [[SigW regulon]]== | ==The [[SigW regulon]]== | ||
| − | '''Additional | + | '''Additional publications:''' {{PubMed|23155385}} |
<pubmed>9683469,16629676,17675383,11866510,9987136,10960106, </pubmed> | <pubmed>9683469,16629676,17675383,11866510,9987136,10960106, </pubmed> | ||
| + | |||
==Other original publications== | ==Other original publications== | ||
'''Additional original publications:''' {{PubMed|20817771,21542858,22178969}} | '''Additional original publications:''' {{PubMed|20817771,21542858,22178969}} | ||
Revision as of 08:56, 23 November 2012
- Description: RNA polymerase ECF-type sigma factor SigW, activated by alkaline shock and by polymyxin B, vancomycin, cephalosporin C, D-cycloserine, and triton X-100
| Gene name | sigW |
| Synonyms | ybbL |
| Essential | no |
| Product | RNA polymerase ECF-type sigma factor SigW |
| Function | resistance against SdpC that functions in detoxification and/or production of antimicrobial compounds |
| Gene expression levels in SubtiExpress: sigW | |
| Interactions involving this protein in SubtInteract: SigW | |
| MW, pI | 21 kDa, 8.614 |
| Gene length, protein length | 561 bp, 187 aa |
| Immediate neighbours | trnSL-Gln2, ybbM |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 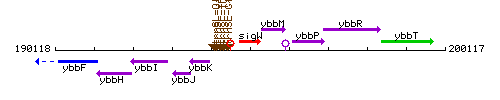
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed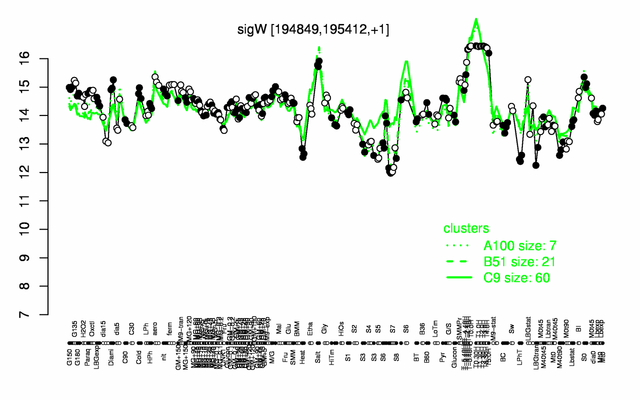
| |
Contents
Categories containing this gene/protein
transcription, sigma factors and their control, cell envelope stress proteins (controlled by SigM, V, W, X, Y)
This gene is a member of the following regulons
The SigW regulon
The gene
Basic information
- Locus tag: BSU01730
Phenotypes of a mutant
- sensitive to ß-lactam antibiotics such as cefuroxime and to fosfomycin PubMed
- Inactivation of alsR, sigD and sigW increases competitive fitness of Bacillus subtilis under non-sporulating conditions PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- A mutation was found in this gene after evolution under relaxed selection for sporulation PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ECF subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity: RsiW acts as antagonist of SigW (anti-SigW)
Database entries
- Structure:
- UniProt: Q45585
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
John Helmann, Cornell University, USA Homepage
Thomas Wiegert, University of Bayreuth, Germany Homepage
Your additional remarks
References
Reviews
Additional reviews: PubMed
The SigW regulon
Additional publications: PubMed
Other original publications
Additional original publications: PubMed