Difference between revisions of "SinR"
| Line 39: | Line 39: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| Line 64: | Line 61: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| − | + | * the mutation suppresses the galactose toxicity to a'' [[galE]]'' mutant {{PubMed|22893383}} | |
=== Database entries === | === Database entries === | ||
| Line 178: | Line 175: | ||
==Original publications== | ==Original publications== | ||
'''Additonal publications:''' {{PubMed|21326214,21708175}} | '''Additonal publications:''' {{PubMed|21326214,21708175}} | ||
| + | <pubmed> 22893383 </pubmed> | ||
<big>''Lehnik-Habrink M, Schaffer M, Mäder U, Diethmaier C, Herzberg C, Stülke J'' </big> | <big>''Lehnik-Habrink M, Schaffer M, Mäder U, Diethmaier C, Herzberg C, Stülke J'' </big> | ||
<big>'''RNA processing in ''Bacillus subtilis'': identification of targets of the essential RNase Y.''' </big> | <big>'''RNA processing in ''Bacillus subtilis'': identification of targets of the essential RNase Y.''' </big> | ||
Revision as of 14:13, 16 August 2012
- Description: transcriptional regulator of post-exponential-phase responses genes
| Gene name | sinR |
| Synonyms | sin, flaD |
| Essential | no |
| Product | transcriptional regulator of post-exponential-phase responses genes |
| Function | control of biofilm formation |
| Gene expression levels in SubtiExpress: sinR | |
| Interactions involving this protein in SubtInteract: SinR | |
| Metabolic function and regulation of this protein in SubtiPathways: Biofilm, Central C-metabolism, Protein secretion | |
| MW, pI | 12 kDa, 7.177 |
| Gene length, protein length | 333 bp, 111 aa |
| Immediate neighbours | sinI, tasA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 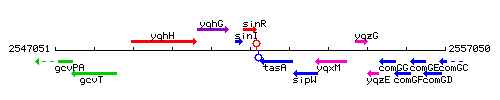
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed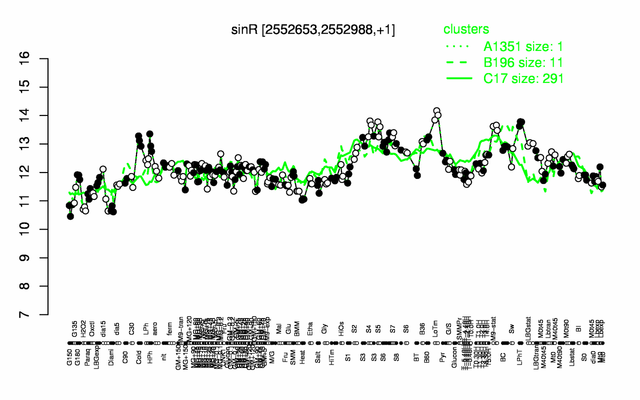
| |
Contents
Categories containing this gene/protein
transcription factors and their control, transition state regulators, biofilm formation
This gene is a member of the following regulons
AbrB regulon, ScoC regulon, Spo0A regulon
The SinR regulon
The gene
Basic information
- Locus tag: BSU24610
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: transcription repressor of biofilm genes, acts as co-repressor for SlrR PubMed
- Protein family:
- Paralogous protein(s): SlrR
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P06533
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: TMB079 sinR::spec, GP736 (tetR), available in Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Modelling of the SinI/SinR switch
Original publications
Additonal publications: PubMed
Lehnik-Habrink M, Schaffer M, Mäder U, Diethmaier C, Herzberg C, Stülke J RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y. Mol Microbiol. 2011 81(6): 1459-1473. PubMed:21815947
Yunrong Chai, Roberto Kolter, Richard Losick
Reversal of an epigenetic switch governing cell chaining in Bacillus subtilis by protein instability.
Mol Microbiol: 2010, 78(1);218-29
[PubMed:20923420]
[WorldCat.org]
[DOI]
(I p)
Yunrong Chai, Thomas Norman, Roberto Kolter, Richard Losick
An epigenetic switch governing daughter cell separation in Bacillus subtilis.
Genes Dev: 2010, 24(8);754-65
[PubMed:20351052]
[WorldCat.org]
[DOI]
(I p)
Prashant Kodgire, K Krishnamurthy Rao
A dual mode of regulation of flgM by ScoC in Bacillus subtilis.
Can J Microbiol: 2009, 55(8);983-9
[PubMed:19898538]
[WorldCat.org]
[DOI]
(I p)
Yunrong Chai, Roberto Kolter, Richard Losick
Paralogous antirepressors acting on the master regulator for biofilm formation in Bacillus subtilis.
Mol Microbiol: 2009, 74(4);876-87
[PubMed:19788541]
[WorldCat.org]
[DOI]
(I p)
Yunrong Chai, Roberto Kolter, Richard Losick
A widely conserved gene cluster required for lactate utilization in Bacillus subtilis and its involvement in biofilm formation.
J Bacteriol: 2009, 191(8);2423-30
[PubMed:19201793]
[WorldCat.org]
[DOI]
(I p)
Frances Chu, Daniel B Kearns, Anna McLoon, Yunrong Chai, Roberto Kolter, Richard Losick
A novel regulatory protein governing biofilm formation in Bacillus subtilis.
Mol Microbiol: 2008, 68(5);1117-27
[PubMed:18430133]
[WorldCat.org]
[DOI]
(I p)
Yunrong Chai, Frances Chu, Roberto Kolter, Richard Losick
Bistability and biofilm formation in Bacillus subtilis.
Mol Microbiol: 2008, 67(2);254-63
[PubMed:18047568]
[WorldCat.org]
[DOI]
(P p)
Prashant Kodgire, Madhulika Dixit, K Krishnamurthy Rao
ScoC and SinR negatively regulate epr by corepression in Bacillus subtilis.
J Bacteriol: 2006, 188(17);6425-8
[PubMed:16923912]
[WorldCat.org]
[DOI]
(P p)
Steven S Branda, Frances Chu, Daniel B Kearns, Richard Losick, Roberto Kolter
A major protein component of the Bacillus subtilis biofilm matrix.
Mol Microbiol: 2006, 59(4);1229-38
[PubMed:16430696]
[WorldCat.org]
[DOI]
(P p)
Frances Chu, Daniel B Kearns, Steven S Branda, Roberto Kolter, Richard Losick
Targets of the master regulator of biofilm formation in Bacillus subtilis.
Mol Microbiol: 2006, 59(4);1216-28
[PubMed:16430695]
[WorldCat.org]
[DOI]
(P p)
Daniel B Kearns, Frances Chu, Steven S Branda, Roberto Kolter, Richard Losick
A master regulator for biofilm formation by Bacillus subtilis.
Mol Microbiol: 2005, 55(3);739-49
[PubMed:15661000]
[WorldCat.org]
[DOI]
(P p)
Alejandro Sánchez, Jorge Olmos
Bacillus subtilis transcriptional regulators interaction.
Biotechnol Lett: 2004, 26(5);403-7
[PubMed:15104138]
[WorldCat.org]
[DOI]
(P p)
Sasha H Shafikhani, Ines Mandic-Mulec, Mark A Strauch, Issar Smith, Terrance Leighton
Postexponential regulation of sin operon expression in Bacillus subtilis.
J Bacteriol: 2002, 184(2);564-71
[PubMed:11751836]
[WorldCat.org]
[DOI]
(P p)
D J Scott, S Leejeerajumnean, J A Brannigan, R J Lewis, A J Wilkinson, J G Hoggett
Quaternary re-arrangement analysed by spectral enhancement: the interaction of a sporulation repressor with its antagonist.
J Mol Biol: 1999, 293(5);997-1004
[PubMed:10547280]
[WorldCat.org]
[DOI]
(P p)
R J Lewis, J A Brannigan, W A Offen, I Smith, A J Wilkinson
An evolutionary link between sporulation and prophage induction in the structure of a repressor:anti-repressor complex.
J Mol Biol: 1998, 283(5);907-12
[PubMed:9799632]
[WorldCat.org]
[DOI]
(P p)
M A Cervin, R J Lewis, J A Brannigan, G B Spiegelman
The Bacillus subtilis regulator SinR inhibits spoIIG promoter transcription in vitro without displacing RNA polymerase.
Nucleic Acids Res: 1998, 26(16);3806-12
[PubMed:9685500]
[WorldCat.org]
[DOI]
(P p)
K Fredrick, J D Helmann
FlgM is a primary regulator of sigmaD activity, and its absence restores motility to a sinR mutant.
J Bacteriol: 1996, 178(23);7010-3
[PubMed:8955328]
[WorldCat.org]
[DOI]
(P p)
M H Rashid, J Sekiguchi
flaD (sinR) mutations affect SigD-dependent functions at multiple points in Bacillus subtilis.
J Bacteriol: 1996, 178(22);6640-3
[PubMed:8932324]
[WorldCat.org]
[DOI]
(P p)
J Hahn, A Luttinger, D Dubnau
Regulatory inputs for the synthesis of ComK, the competence transcription factor of Bacillus subtilis.
Mol Microbiol: 1996, 21(4);763-75
[PubMed:8878039]
[WorldCat.org]
[DOI]
(P p)
P Margot, V Lazarevic, D Karamata
Effect of the SinR protein on the expression of the Bacillus subtilis 168 lytABC operon.
Microb Drug Resist: 1996, 2(1);119-21
[PubMed:9158733]
[WorldCat.org]
[DOI]
(P p)
M A Strauch
In vitro binding affinity of the Bacillus subtilis AbrB protein to six different DNA target regions.
J Bacteriol: 1995, 177(15);4532-6
[PubMed:7635837]
[WorldCat.org]
[DOI]
(P p)
U Bai, I Mandic-Mulec, I Smith
SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction.
Genes Dev: 1993, 7(1);139-48
[PubMed:8422983]
[WorldCat.org]
[DOI]
(P p)
P T Kallio, J E Fagelson, J A Hoch, M A Strauch
The transition state regulator Hpr of Bacillus subtilis is a DNA-binding protein.
J Biol Chem: 1991, 266(20);13411-7
[PubMed:1906467]
[WorldCat.org]
(P p)
N K Gaur, K Cabane, I Smith
Structure and expression of the Bacillus subtilis sin operon.
J Bacteriol: 1988, 170(3);1046-53
[PubMed:3125149]
[WorldCat.org]
[DOI]
(P p)