Difference between revisions of "RsiW"
(→Categories containing this gene/protein) |
(→Extended information on the protein) |
||
| Line 85: | Line 85: | ||
* '''Effectors of protein activity:''' degraded by [[RasP]] and [[PrsW]] under conditions of alkali shock or in the presence of antimicrobial peptides, respectively. This results in the release of [[SigW]] | * '''Effectors of protein activity:''' degraded by [[RasP]] and [[PrsW]] under conditions of alkali shock or in the presence of antimicrobial peptides, respectively. This results in the release of [[SigW]] | ||
| − | * '''Interactions:''' [[PrsW]]-[[RsiW]] (proteolysis) {{PubMed|17020587}} | + | * '''[[SubtInteract|Interactions]]:''' |
| + | ** [[PrsW]]-[[RsiW]] (proteolysis) {{PubMed|17020587}} | ||
| + | ** [[RasP]]-[[RsiW]] (proteolysis), [[RsiW]]-[[SigW]] | ||
| − | * '''Localization:''' membrane (according to Swiss-Prot) | + | * '''[[Localization]]:''' membrane (according to Swiss-Prot) |
=== Database entries === | === Database entries === | ||
Revision as of 09:14, 2 November 2011
- Description: anti-SigW
| Gene name | rsiW |
| Synonyms | ybbM |
| Essential | no |
| Product | anti-SigW |
| Function | control of SigW activity |
| Interactions involving this protein in SubtInteract: RsiW | |
| MW, pI | 23 kDa, 6.646 |
| Gene length, protein length | 624 bp, 208 aa |
| Immediate neighbours | sigW, ybbP |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 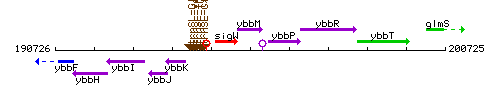
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
sigma factors and their control, cell envelope stress proteins (controlled by SigM, V, W, X, Y), membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU01740
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: anti-sigma-W factor family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity: degraded by RasP and PrsW under conditions of alkali shock or in the presence of antimicrobial peptides, respectively. This results in the release of SigW
- Localization: membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: Q45588
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information: the mRNA is very stable (half-life > 15 min) PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Thomas Wiegert, University of Bayreuth, Germany Homepage
Your additional remarks
References
Reviews=
Original Publications
Janine Heinrich, Kerstin Hein, Thomas Wiegert
Two proteolytic modules are involved in regulated intramembrane proteolysis of Bacillus subtilis RsiW.
Mol Microbiol: 2009, 74(6);1412-26
[PubMed:19889088]
[WorldCat.org]
[DOI]
(I p)
Janine Heinrich, Tuula Lundén, Vesa P Kontinen, Thomas Wiegert
The Bacillus subtilis ABC transporter EcsAB influences intramembrane proteolysis through RasP.
Microbiology (Reading): 2008, 154(Pt 7);1989-1997
[PubMed:18599827]
[WorldCat.org]
[DOI]
(P p)
Janine Heinrich, Thomas Wiegert
YpdC determines site-1 degradation in regulated intramembrane proteolysis of the RsiW anti-sigma factor of Bacillus subtilis.
Mol Microbiol: 2006, 62(2);566-79
[PubMed:17020587]
[WorldCat.org]
[DOI]
(P p)
Stephan Zellmeier, Wolfgang Schumann, Thomas Wiegert
Involvement of Clp protease activity in modulating the Bacillus subtilissigmaw stress response.
Mol Microbiol: 2006, 61(6);1569-82
[PubMed:16899079]
[WorldCat.org]
[DOI]
(P p)
Craig D Ellermeier, Richard Losick
Evidence for a novel protease governing regulated intramembrane proteolysis and resistance to antimicrobial peptides in Bacillus subtilis.
Genes Dev: 2006, 20(14);1911-22
[PubMed:16816000]
[WorldCat.org]
[DOI]
(P p)
Susanne Schöbel, Stephan Zellmeier, Wolfgang Schumann, Thomas Wiegert
The Bacillus subtilis sigmaW anti-sigma factor RsiW is degraded by intramembrane proteolysis through YluC.
Mol Microbiol: 2004, 52(4);1091-105
[PubMed:15130127]
[WorldCat.org]
[DOI]
(P p)
Mika Yoshimura, Kei Asai, Yoshito Sadaie, Hirofumi Yoshikawa
Interaction of Bacillus subtilis extracytoplasmic function (ECF) sigma factors with the N-terminal regions of their potential anti-sigma factors.
Microbiology (Reading): 2004, 150(Pt 3);591-599
[PubMed:14993308]
[WorldCat.org]
[DOI]
(P p)
G Hambraeus, C von Wachenfeldt, L Hederstedt
Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs.
Mol Genet Genomics: 2003, 269(5);706-14
[PubMed:12884008]
[WorldCat.org]
[DOI]
(P p)
Min Cao, Tao Wang, Rick Ye, John D Helmann
Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis sigma(W) and sigma(M) regulons.
Mol Microbiol: 2002, 45(5);1267-76
[PubMed:12207695]
[WorldCat.org]
[DOI]
(P p)
Qiang Qian, Chien Y Lee, John D Helmann, Mark A Strauch
AbrB is a regulator of the sigma(W) regulon in Bacillus subtilis.
FEMS Microbiol Lett: 2002, 211(2);219-23
[PubMed:12076816]
[WorldCat.org]
[DOI]
(P p)