Difference between revisions of "CitB"
(→Original publications) |
(→Expression and regulation) |
||
| Line 118: | Line 118: | ||
** repressed during growth in the presence of branched chain amino acids ([[CodY]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/12591885 PubMed] | ** repressed during growth in the presence of branched chain amino acids ([[CodY]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/12591885 PubMed] | ||
** repressed in the presence of glucose and glutamate ([[CcpC]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/10656796 PubMed] | ** repressed in the presence of glucose and glutamate ([[CcpC]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/10656796 PubMed] | ||
| − | ** expressed upon transition into the stationary phase ([[AbrB]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/12591885 PubMed] | + | ** expressed upon transition into the stationary phase ([[AbrB]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/12591885 PubMed], probably indirect negative regulation by [[AbrB]] {{PubMed|20817675}} |
** repressed by glucose (3.7-fold) ([[CcpA]]) [http://www.ncbi.nlm.nih.gov/pubmed/12850135 PubMed] | ** repressed by glucose (3.7-fold) ([[CcpA]]) [http://www.ncbi.nlm.nih.gov/pubmed/12850135 PubMed] | ||
** repression by glucose + arginine ([[CcpC]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/16395550 PubMed] | ** repression by glucose + arginine ([[CcpC]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/16395550 PubMed] | ||
| Line 126: | Line 126: | ||
** [[CodY]]: transcription repression [http://www.ncbi.nlm.nih.gov/sites/entrez/12591885 PubMed] | ** [[CodY]]: transcription repression [http://www.ncbi.nlm.nih.gov/sites/entrez/12591885 PubMed] | ||
** [[CcpC]]: transcription repression (molecular inducer: citrate) [http://www.ncbi.nlm.nih.gov/sites/entrez/10656796 PubMed1] [http://www.ncbi.nlm.nih.gov/sites/entrez/12100558 PubMed2] | ** [[CcpC]]: transcription repression (molecular inducer: citrate) [http://www.ncbi.nlm.nih.gov/sites/entrez/10656796 PubMed1] [http://www.ncbi.nlm.nih.gov/sites/entrez/12100558 PubMed2] | ||
| − | |||
** [[CcpA]]: transcription repression [http://www.ncbi.nlm.nih.gov/sites/entrez/12100558 PubMed] | ** [[CcpA]]: transcription repression [http://www.ncbi.nlm.nih.gov/sites/entrez/12100558 PubMed] | ||
** [[FsrA]]: translational repression [http://www.ncbi.nlm.nih.gov/pubmed/18697947 PubMed] | ** [[FsrA]]: translational repression [http://www.ncbi.nlm.nih.gov/pubmed/18697947 PubMed] | ||
Revision as of 17:10, 30 May 2011
- Description: trigger enzyme: aconitase and RNA binding protein
| Gene name | citB |
| Synonyms | |
| Essential | no |
| Product | trigger enzyme: aconitate hydratase (aconitase) |
| Function | TCA cycle |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 99 kDa, 4.903 |
| Gene length, protein length | 2727 bp, 909 aa |
| Immediate neighbours | sspO, yneN |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 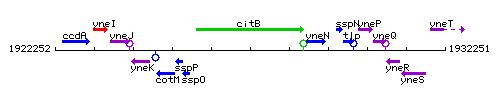
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
carbon core metabolism, trigger enzyme, RNA binding regulators
This gene is a member of the following regulons
AbrB regulon, CcpC regulon, CodY regulon, FsrA regulon
The CitB regulon: feuA-feuB-feuC-ybbA
The gene
Basic information
- Locus tag: BSU18000
Phenotypes of a mutant
glutamate auxotrophy and a defect in sporulation PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Citrate = isocitrate (according to Swiss-Prot)
- Binding to iron responsive elements (IRE RNA) in the absence of the FeS cluster PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s): FeS cluster
- Effectors of protein activity:
- Localization:
Database entries
- Structure: 1L5J (E. coli)
- UniProt: P09339
- KEGG entry: [3]
- E.C. number: 4.2.1.3
Additional information
- B. subtilis aconitase is both an enzyme and an RNA binding protein (moonlighting protein) PubMed
- extensive information on the structure and enzymatic properties of CitB can be found at Proteopedia
Expression and regulation
- Regulation:
- repressed during growth in the presence of branched chain amino acids (CodY) PubMed
- repressed in the presence of glucose and glutamate (CcpC) PubMed
- expressed upon transition into the stationary phase (AbrB) PubMed, probably indirect negative regulation by AbrB PubMed
- repressed by glucose (3.7-fold) (CcpA) PubMed
- repression by glucose + arginine (CcpC) PubMed
- less expressed under conditions of extreme iron limitation (FsrA) PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: GP683 (erm), available in Stülke lab
- Expression vector:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody: available in Linc Sonenshein lab
Labs working on this gene/protein
Linc Sonenshein, Tufts University, Boston, MA, USA Homepage
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References
Reviews
Original publications