Difference between revisions of "PolY2"
| (31 intermediate revisions by 6 users not shown) | |||
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || UV-targeted mutagenesis | |style="background:#ABCDEF;" align="center"|'''Function''' || UV-targeted mutagenesis | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU23710 polY2] | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://subtiwiki.uni-goettingen.de/interact/ ''Subt''Interact]''': [http://subtiwiki.uni-goettingen.de/interact/index.php?protein=PolY2 PolY2] | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 45 kDa, 9.532 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 45 kDa, 9.532 | ||
| Line 20: | Line 24: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yqjX]]'', ''[[yqzH]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yqjX]]'', ''[[yqzH]]'' | ||
|- | |- | ||
| − | | | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU23710 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU23710 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU23710 DNA_with_flanks] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:yqjW_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:yqjW_context.gif]] | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
| + | |- | ||
| + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=polYB_2464562_2465800_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:polY2_expression.png|500px|link=http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU23710]] | ||
|- | |- | ||
|} | |} | ||
__TOC__ | __TOC__ | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/> | ||
| − | + | = [[Categories]] containing this gene/protein = | |
| + | {{SubtiWiki category|[[DNA repair/ recombination]]}} | ||
| + | |||
| + | = This gene is a member of the following [[regulons]] = | ||
| + | {{SubtiWiki regulon|[[LexA regulon]]}} | ||
=The gene= | =The gene= | ||
| Line 35: | Line 49: | ||
=== Basic information === | === Basic information === | ||
| − | * '''Locus tag:''' | + | * '''Locus tag:''' BSU23710 |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU23710&redirect=T BSU23710] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/yqjWX.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/yqjWX.html] | ||
| Line 46: | Line 61: | ||
=== Additional information=== | === Additional information=== | ||
| + | |||
| + | |||
| Line 56: | Line 73: | ||
* '''Protein family:''' DNA polymerase type-Y family (according to Swiss-Prot) | * '''Protein family:''' DNA polymerase type-Y family (according to Swiss-Prot) | ||
| − | * '''Paralogous protein(s):''' | + | * '''Paralogous protein(s):''' [[PolY1]], [[UvrX]], [[YozK]]-[[YobH]] |
=== Extended information on the protein === | === Extended information on the protein === | ||
| Line 70: | Line 87: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' | + | * '''[[SubtInteract|Interactions]]:''' |
| + | ** [[PolY2]]-[[DnaN]] (required for untargeted mutagenesis) {{PubMed|15469515}} | ||
| + | ** [[PolA]]-[[PolY2]] {{PubMed|16045613}} | ||
| − | * '''Localization:''' | + | * '''[[Localization]]:''' |
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU23710&redirect=T BSU23710] | ||
* '''Structure:''' | * '''Structure:''' | ||
| − | * ''' | + | * '''UniProt:''' [http://www.uniprot.org/uniprot/P54560 P54560] |
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU23710] |
* '''E.C. number:''' | * '''E.C. number:''' | ||
| Line 88: | Line 108: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' | + | * '''Operon:''' ''[[polY2]]-[[yqjX]]'' (according to [http://dbtbs.hgc.jp/COG/prom/yqjWX.html DBTBS]) |
| + | |||
| + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=polYB_2464562_2465800_-1 polY2] {{PubMed|22383849}} | ||
* '''[[Sigma factor]]:''' | * '''[[Sigma factor]]:''' | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| + | ** induced upon DNA damage ([[LexA]]) {{PubMed|15469515,16267290}} | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| + | ** [[LexA]]: transcription repression {{PubMed|15469515,16267290}} | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
=Biological materials = | =Biological materials = | ||
| − | * '''Mutant:''' | + | * '''Mutant:''' GP1505 (''polY2''::''kan''), available in [[Stülke]] lab |
* '''Expression vector:''' | * '''Expression vector:''' | ||
| Line 117: | Line 141: | ||
=References= | =References= | ||
| + | ==Reviews== | ||
| + | <pubmed> 22933559 </pubmed> | ||
| + | == Original publications == | ||
| + | <pubmed>16267290, 12644484 15469515 16045613 19924481 23686288 </pubmed> | ||
| − | + | [[Category:Protein-coding genes]] | |
Latest revision as of 14:08, 2 April 2014
- Description: translesion synthesis (TLS-) DNA polymerase Y2
| Gene name | polY2 |
| Synonyms | yqjW |
| Essential | no |
| Product | translesion synthesis (TLS-) DNA polymerase Y2 |
| Function | UV-targeted mutagenesis |
| Gene expression levels in SubtiExpress: polY2 | |
| Interactions involving this protein in SubtInteract: PolY2 | |
| MW, pI | 45 kDa, 9.532 |
| Gene length, protein length | 1236 bp, 412 aa |
| Immediate neighbours | yqjX, yqzH |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 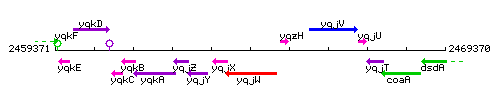
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed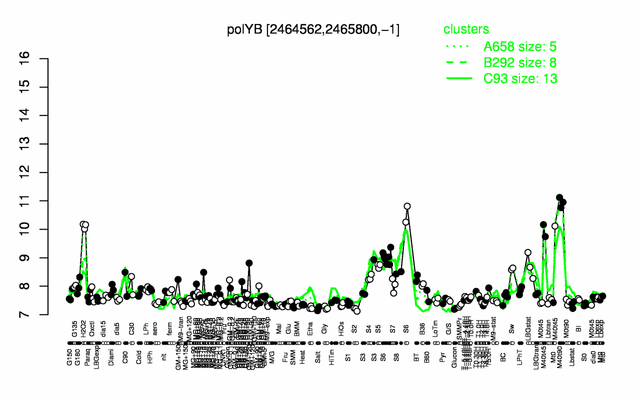
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU23710
Phenotypes of a mutant
Database entries
- BsubCyc: BSU23710
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Deoxynucleoside triphosphate + DNA(n) = diphosphate + DNA(n+1) (according to Swiss-Prot)
- Protein family: DNA polymerase type-Y family (according to Swiss-Prot)
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU23710
- Structure:
- UniProt: P54560
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant: GP1505 (polY2::kan), available in Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications