Difference between revisions of "Nfo"
(→Phenotypes of a mutant) |
|||
| (5 intermediate revisions by 3 users not shown) | |||
| Line 53: | Line 53: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| − | + | * an ''[[exoA]] [[nfo]]'' double mutant is impaired in germination and spore outgrowth due to the accumulation of DNA lesions, this can be rescued by inactivation of ''[[disA]]'' {{PubMed|24244006}} | |
| + | * an ''[[exoA]] [[nfo]]'' double mutant is sensitive to radiation {{PubMed|24123749}} | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU25130&redirect=T BSU25130] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/yqfSU.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/yqfSU.html] | ||
| Line 90: | Line 92: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU25130&redirect=T BSU25130] | ||
* '''Structure:''' | * '''Structure:''' | ||
| Line 115: | Line 118: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
The gene ''[[yqfT]]'' is located between ''[[nfo]]'' and ''[[yqfU]]'', but is transcribed in the opposite direction. | The gene ''[[yqfT]]'' is located between ''[[nfo]]'' and ''[[yqfU]]'', but is transcribed in the opposite direction. | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 262 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (complex medium with amino acids, without glucose): 659 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 612 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 355 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 628 {{PubMed|21395229}} | ||
=Biological materials = | =Biological materials = | ||
| − | |||
* '''Mutant:''' | * '''Mutant:''' | ||
** available in [[Mario Pedraza-Reyes]]' lab | ** available in [[Mario Pedraza-Reyes]]' lab | ||
| Line 140: | Line 147: | ||
<pubmed> 22933559 </pubmed> | <pubmed> 22933559 </pubmed> | ||
== Original publications == | == Original publications == | ||
| − | <pubmed>24244006,18203828,16237020,12949090, 19930460,12486072, 21441501 24123749 </pubmed> | + | <pubmed>24244006,18203828,16237020,12949090, 19930460,12486072, 21441501 24123749 24123749 24914186 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Latest revision as of 08:02, 11 June 2014
- Description: type IV apurinic/apyrimidinic endonuclease
| Gene name | nfo |
| Synonyms | yqfS |
| Essential | no |
| Product | type IV apurinic/apyrimidinic endonuclease |
| Function | repair of oxidative DNA damage in spores |
| Gene expression levels in SubtiExpress: nfo | |
| MW, pI | 32 kDa, 5.371 |
| Gene length, protein length | 891 bp, 297 aa |
| Immediate neighbours | yqfT, cshB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 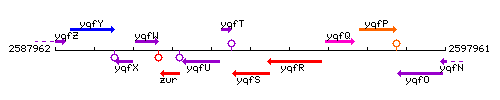
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
DNA repair/ recombination, sporulation proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU25130
Phenotypes of a mutant
- an exoA nfo double mutant is impaired in germination and spore outgrowth due to the accumulation of DNA lesions, this can be rescued by inactivation of disA PubMed
- an exoA nfo double mutant is sensitive to radiation PubMed
Database entries
- BsubCyc: BSU25130
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- Nfo is functionally redundant with ExoA
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Endonucleolytic cleavage to 5'-phosphooligonucleotide end-products (according to Swiss-Prot)
- Protein family: AP endonuclease 2 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU25130
- Structure:
- UniProt: P54476
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
The gene yqfT is located between nfo and yqfU, but is transcribed in the opposite direction.
- number of protein molecules per cell (minimal medium with glucose and ammonium): 262 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 659 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 612 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 355 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 628 PubMed
Biological materials
- Mutant:
- available in Mario Pedraza-Reyes' lab
- GP899 (nfo::kan) and GP1502 (nfo::cat), available in Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications