Difference between revisions of "Nfo"
(→References) |
|||
| (21 intermediate revisions by 5 users not shown) | |||
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || repair of oxidative DNA damage in spores | |style="background:#ABCDEF;" align="center"|'''Function''' || repair of oxidative DNA damage in spores | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU25130 nfo] | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 32 kDa, 5.371 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 32 kDa, 5.371 | ||
| Line 20: | Line 22: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yqfT]]'', ''[[cshB]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yqfT]]'', ''[[cshB]]'' | ||
|- | |- | ||
| − | | | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU25130 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU25130 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU25130 DNA_with_flanks] |
|- | |- | ||
|- | |- | ||
| Line 26: | Line 28: | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:yqfS_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:yqfS_context.gif]] | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
| + | |- | ||
| + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=nfo_2593281_2594174_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:nfo_expression.png|500px|link=http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU25130]] | ||
|- | |- | ||
|} | |} | ||
__TOC__ | __TOC__ | ||
| − | |||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/> | ||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
| Line 47: | Line 53: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| + | * an ''[[exoA]] [[nfo]]'' double mutant is impaired in germination and spore outgrowth due to the accumulation of DNA lesions, this can be rescued by inactivation of ''[[disA]]'' {{PubMed|24244006}} | ||
| + | * an ''[[exoA]] [[nfo]]'' double mutant is sensitive to radiation {{PubMed|24123749}} | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU25130&redirect=T BSU25130] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/yqfSU.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/yqfSU.html] | ||
| Line 55: | Line 64: | ||
=== Additional information=== | === Additional information=== | ||
| − | + | * Nfo is functionally redundant with [[ExoA]] | |
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
| Line 81: | Line 87: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' | + | * '''[[SubtInteract|Interactions]]:''' |
| − | * '''Localization:''' | + | * '''[[Localization]]:''' |
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU25130&redirect=T BSU25130] | ||
* '''Structure:''' | * '''Structure:''' | ||
| Line 100: | Line 107: | ||
* '''Operon:''' ''[[nfo]]-[[yqfU]]'' {{PubMed|12486072}} | * '''Operon:''' ''[[nfo]]-[[yqfU]]'' {{PubMed|12486072}} | ||
| − | * '''[ | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=nfo_2593281_2594174_-1 nfo] {{PubMed|22383849}} |
| + | |||
| + | * '''Sigma factor:''' [[SigG]] {{PubMed|12486072}} | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 109: | Line 118: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
The gene ''[[yqfT]]'' is located between ''[[nfo]]'' and ''[[yqfU]]'', but is transcribed in the opposite direction. | The gene ''[[yqfT]]'' is located between ''[[nfo]]'' and ''[[yqfU]]'', but is transcribed in the opposite direction. | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 262 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (complex medium with amino acids, without glucose): 659 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 612 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 355 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 628 {{PubMed|21395229}} | ||
=Biological materials = | =Biological materials = | ||
| − | + | * '''Mutant:''' | |
| − | * '''Mutant:''' available in [[Mario Pedraza-Reyes]]' lab | + | ** available in [[Mario Pedraza-Reyes]]' lab |
| + | ** GP899 (''nfo''::''kan'') and GP1502 (''nfo''::''cat''), available in [[Stülke]] lab | ||
* '''Expression vector:''' | * '''Expression vector:''' | ||
| Line 129: | Line 144: | ||
=References= | =References= | ||
| − | <pubmed>18203828,16237020,12949090, 19930460,12486072, 21441501 </pubmed> | + | ==Reviews== |
| + | <pubmed> 22933559 </pubmed> | ||
| + | == Original publications == | ||
| + | <pubmed>24244006,18203828,16237020,12949090, 19930460,12486072, 21441501 24123749 24123749 24914186 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Latest revision as of 08:02, 11 June 2014
- Description: type IV apurinic/apyrimidinic endonuclease
| Gene name | nfo |
| Synonyms | yqfS |
| Essential | no |
| Product | type IV apurinic/apyrimidinic endonuclease |
| Function | repair of oxidative DNA damage in spores |
| Gene expression levels in SubtiExpress: nfo | |
| MW, pI | 32 kDa, 5.371 |
| Gene length, protein length | 891 bp, 297 aa |
| Immediate neighbours | yqfT, cshB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed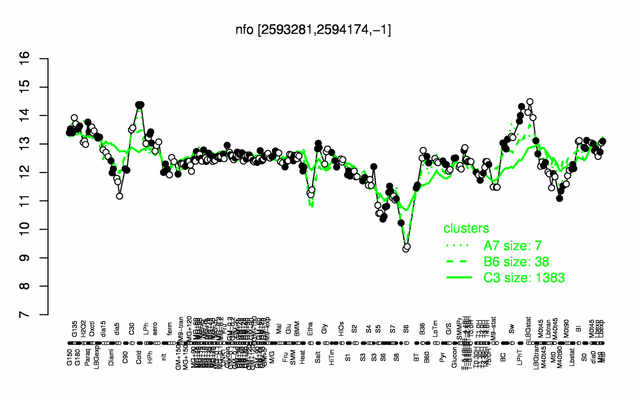
| |
Contents
Categories containing this gene/protein
DNA repair/ recombination, sporulation proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU25130
Phenotypes of a mutant
- an exoA nfo double mutant is impaired in germination and spore outgrowth due to the accumulation of DNA lesions, this can be rescued by inactivation of disA PubMed
- an exoA nfo double mutant is sensitive to radiation PubMed
Database entries
- BsubCyc: BSU25130
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- Nfo is functionally redundant with ExoA
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Endonucleolytic cleavage to 5'-phosphooligonucleotide end-products (according to Swiss-Prot)
- Protein family: AP endonuclease 2 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU25130
- Structure:
- UniProt: P54476
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
The gene yqfT is located between nfo and yqfU, but is transcribed in the opposite direction.
- number of protein molecules per cell (minimal medium with glucose and ammonium): 262 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 659 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 612 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 355 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 628 PubMed
Biological materials
- Mutant:
- available in Mario Pedraza-Reyes' lab
- GP899 (nfo::kan) and GP1502 (nfo::cat), available in Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications
Rocío del Carmen Barajas-Ornelas, Fernando H Ramírez-Guadiana, Rafael Juárez-Godínez, Victor M Ayala-García, Eduardo A Robleto, Ronald E Yasbin, Mario Pedraza-Reyes
Error-prone processing of apurinic/apyrimidinic (AP) sites by PolX underlies a novel mechanism that promotes adaptive mutagenesis in Bacillus subtilis.
J Bacteriol: 2014, 196(16);3012-22
[PubMed:24914186]
[WorldCat.org]
[DOI]
(I p)
Silvia S Campos, Juan R Ibarra-Rodriguez, Rocío C Barajas-Ornelas, Fernando H Ramírez-Guadiana, Armando Obregón-Herrera, Peter Setlow, Mario Pedraza-Reyes
Interaction of apurinic/apyrimidinic endonucleases Nfo and ExoA with the DNA integrity scanning protein DisA in the processing of oxidative DNA damage during Bacillus subtilis spore outgrowth.
J Bacteriol: 2014, 196(3);568-78
[PubMed:24244006]
[WorldCat.org]
[DOI]
(I p)
Ralf Moeller, Marina Raguse, Günther Reitz, Ryuichi Okayasu, Zuofeng Li, Stuart Klein, Peter Setlow, Wayne L Nicholson
Resistance of Bacillus subtilis spore DNA to lethal ionizing radiation damage relies primarily on spore core components and DNA repair, with minor effects of oxygen radical detoxification.
Appl Environ Microbiol: 2014, 80(1);104-9
[PubMed:24123749]
[WorldCat.org]
[DOI]
(I p)
Ralf Moeller, Peter Setlow, Mario Pedraza-Reyes, Ryuichi Okayasu, Günther Reitz, Wayne L Nicholson
Role of the Nfo and ExoA apurinic/apyrimidinic endonucleases in radiation resistance and radiation-induced mutagenesis of Bacillus subtilis spores.
J Bacteriol: 2011, 193(11);2875-9
[PubMed:21441501]
[WorldCat.org]
[DOI]
(I p)
Marcelo Barraza-Salas, Juan R Ibarra-Rodríguez, Silvia J Mellado, José M Salas-Pacheco, Peter Setlow, Mario Pedraza-Reyes
Effects of forespore-specific overexpression of apurinic/apyrimidinic endonuclease Nfo on the DNA-damage resistance properties of Bacillus subtilis spores.
FEMS Microbiol Lett: 2010, 302(2);159-65
[PubMed:19930460]
[WorldCat.org]
[DOI]
(I p)
Juan R Ibarra, Alma D Orozco, Juan A Rojas, Karina López, Peter Setlow, Ronald E Yasbin, Mario Pedraza-Reyes
Role of the Nfo and ExoA apurinic/apyrimidinic endonucleases in repair of DNA damage during outgrowth of Bacillus subtilis spores.
J Bacteriol: 2008, 190(6);2031-8
[PubMed:18203828]
[WorldCat.org]
[DOI]
(I p)
José M Salas-Pacheco, Barbara Setlow, Peter Setlow, Mario Pedraza-Reyes
Role of the Nfo (YqfS) and ExoA apurinic/apyrimidinic endonucleases in protecting Bacillus subtilis spores from DNA damage.
J Bacteriol: 2005, 187(21);7374-81
[PubMed:16237020]
[WorldCat.org]
[DOI]
(P p)
José M Salas-Pacheco, Norma Urtiz-Estrada, Guadalupe Martínez-Cadena, Ronald E Yasbin, Mario Pedraza-Reyes
YqfS from Bacillus subtilis is a spore protein and a new functional member of the type IV apurinic/apyrimidinic-endonuclease family.
J Bacteriol: 2003, 185(18);5380-90
[PubMed:12949090]
[WorldCat.org]
[DOI]
(P p)
Norma Urtiz-Estrada, José M Salas-Pacheco, Ronald E Yasbin, Mario Pedraza-Reyes
Forespore-specific expression of Bacillus subtilis yqfS, which encodes type IV apurinic/apyrimidinic endonuclease, a component of the base excision repair pathway.
J Bacteriol: 2003, 185(1);340-8
[PubMed:12486072]
[WorldCat.org]
[DOI]
(P p)