Difference between revisions of "CwlS"
| (17 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | * '''Description:''' D,L-endopeptidase, peptidoglycan hydrolase <br/><br/> | + | * '''Description:''' D,L-endopeptidase, peptidoglycan hydrolase for cell separation<br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 12: | Line 12: | ||
|style="background:#ABCDEF;" align="center"| '''Product''' || D,L-endopeptidase, peptidoglycan hydrolase | |style="background:#ABCDEF;" align="center"| '''Product''' || D,L-endopeptidase, peptidoglycan hydrolase | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || cell | + | |style="background:#ABCDEF;" align="center"|'''Function''' || cell separation |
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU19410 cwlS] | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 44 kDa, 10.438 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 44 kDa, 10.438 | ||
| Line 20: | Line 22: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yojM]]'', ''[[yojK]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yojM]]'', ''[[yojK]]'' | ||
|- | |- | ||
| − | | | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU19410 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU19410 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU19410 DNA_with_flanks] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:yojL_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:yojL_context.gif]] | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
| + | |- | ||
| + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=cwlS_2115425_2116669_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:cwlS_expression.png|500px|link=http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU19410]] | ||
|- | |- | ||
|} | |} | ||
__TOC__ | __TOC__ | ||
| − | + | <br/><br/><br/><br/> | |
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
<br/><br/> | <br/><br/> | ||
| Line 36: | Line 42: | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| − | {{SubtiWiki regulon|[[CcpA regulon]]}} | + | {{SubtiWiki regulon|[[Abh regulon]]}}, |
| + | {{SubtiWiki regulon|[[AbrB regulon]]}}, | ||
| + | {{SubtiWiki regulon|[[CcpA regulon]]}}, | ||
| + | {{SubtiWiki regulon|[[SigD regulon]]}}, | ||
| + | {{SubtiWiki regulon|[[SigH regulon]]}}, | ||
=The gene= | =The gene= | ||
| Line 47: | Line 57: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU19410&redirect=T BSU19410] | ||
* '''DBTBS entry:''' no entry | * '''DBTBS entry:''' no entry | ||
| Line 53: | Line 64: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| Line 65: | Line 74: | ||
* '''Protein family:''' Cu-Zn superoxide dismutase family (according to Swiss-Prot) | * '''Protein family:''' Cu-Zn superoxide dismutase family (according to Swiss-Prot) | ||
| − | * '''Paralogous protein(s):''' | + | * '''Paralogous protein(s):''' the C-terminal D,L-endopeptidase domains of [[LytE]], [[LytF]], [[CwlS]], and [[CwlO]] exhibit strong sequence similarity |
=== Extended information on the protein === | === Extended information on the protein === | ||
| Line 71: | Line 80: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
| + | ** contains four N-acetylglucosamine-polymer-binding [[LysM domain]]s {{PubMed|24355088,18430080}} | ||
| + | ** C-terminal D,L-endopeptidase domain {{PubMed|22139507}} | ||
* '''Modification:''' | * '''Modification:''' | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' | + | * '''[[SubtInteract|Interactions]]:''' |
| − | * '''Localization:''' cell membrane (according to Swiss-Prot) | + | * '''[[Localization]]:''' |
| + | ** cell membrane (according to Swiss-Prot) | ||
| + | ** localizes to cell septa and poles {{PubMed|22139507}} | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU19410&redirect=T BSU19410] | ||
* '''Structure:''' | * '''Structure:''' | ||
| Line 99: | Line 113: | ||
* '''Operon:''' | * '''Operon:''' | ||
| − | * '''[[Sigma factor]]:''' | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=cwlS_2115425_2116669_-1 cwlS] {{PubMed|22383849}} |
| + | |||
| + | * '''[[Sigma factor]]:''' [[SigD]], [[SigH]], according to {{PubMed|22139507}} | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 129: | Line 145: | ||
=References= | =References= | ||
| − | + | == Reviews== | |
| − | <pubmed>16855244 | + | <pubmed>18430080</pubmed> |
| − | + | == Original publications == | |
| + | <pubmed>16855244,12850135, 22139507 20817675 24355088</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Latest revision as of 13:55, 2 April 2014
- Description: D,L-endopeptidase, peptidoglycan hydrolase for cell separation
| Gene name | cwlS |
| Synonyms | yojL |
| Essential | no |
| Product | D,L-endopeptidase, peptidoglycan hydrolase |
| Function | cell separation |
| Gene expression levels in SubtiExpress: cwlS | |
| MW, pI | 44 kDa, 10.438 |
| Gene length, protein length | 1242 bp, 414 aa |
| Immediate neighbours | yojM, yojK |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 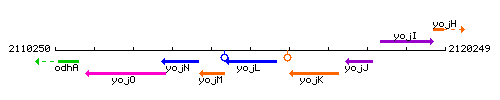
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed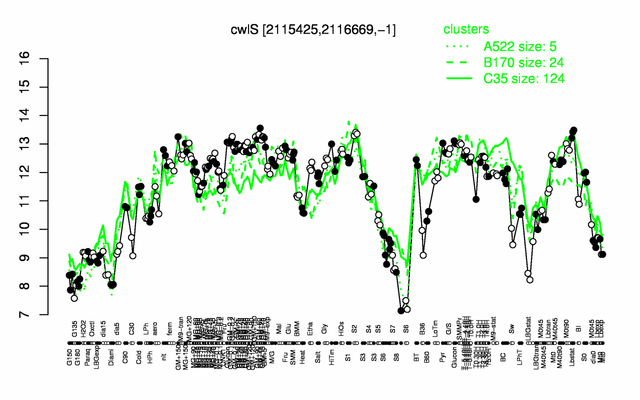
| |
Contents
Categories containing this gene/protein
cell wall degradation/ turnover, membrane proteins
This gene is a member of the following regulons
Abh regulon, AbrB regulon, CcpA regulon, SigD regulon, SigH regulon,
The gene
Basic information
- Locus tag: BSU19410
Phenotypes of a mutant
Database entries
- BsubCyc: BSU19410
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Hydrolysis of gamma-D-glutamyl bonds to the L-terminus (position 7) of meso-diaminopimelic acid (meso-A2pm) in 7-(L-Ala-gamma-D-Glu)-meso-A2pm and 7-(L-Ala-gamma-D-Glu)-7-(D-Ala)-meso-A2pm (according to Swiss-Prot)
- Protein family: Cu-Zn superoxide dismutase family (according to Swiss-Prot)
- Paralogous protein(s): the C-terminal D,L-endopeptidase domains of LytE, LytF, CwlS, and CwlO exhibit strong sequence similarity
Extended information on the protein
- Kinetic information:
- Domains:
- contains four N-acetylglucosamine-polymer-binding LysM domains PubMed
- C-terminal D,L-endopeptidase domain PubMed
- Modification:
- Effectors of protein activity:
- Localization:
- cell membrane (according to Swiss-Prot)
- localizes to cell septa and poles PubMed
Database entries
- BsubCyc: BSU19410
- Structure:
- UniProt: O31852
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor: SigD, SigH, according to PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications
Jaslyn E M M Wong, Husam M A B Alsarraf, Jørn Døvling Kaspersen, Jan Skov Pedersen, Jens Stougaard, Søren Thirup, Mickaël Blaise
Cooperative binding of LysM domains determines the carbohydrate affinity of a bacterial endopeptidase protein.
FEBS J: 2014, 281(4);1196-208
[PubMed:24355088]
[WorldCat.org]
[DOI]
(I p)
Masayuki Hashimoto, Seika Ooiwa, Junichi Sekiguchi
Synthetic lethality of the lytE cwlO genotype in Bacillus subtilis is caused by lack of D,L-endopeptidase activity at the lateral cell wall.
J Bacteriol: 2012, 194(4);796-803
[PubMed:22139507]
[WorldCat.org]
[DOI]
(I p)
Onuma Chumsakul, Hiroki Takahashi, Taku Oshima, Takahiro Hishimoto, Shigehiko Kanaya, Naotake Ogasawara, Shu Ishikawa
Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation.
Nucleic Acids Res: 2011, 39(2);414-28
[PubMed:20817675]
[WorldCat.org]
[DOI]
(I p)
Tatsuya Fukushima, Anahita Afkham, Shin-Ichirou Kurosawa, Taichi Tanabe, Hiroki Yamamoto, Junichi Sekiguchi
A new D,L-endopeptidase gene product, YojL (renamed CwlS), plays a role in cell separation with LytE and LytF in Bacillus subtilis.
J Bacteriol: 2006, 188(15);5541-50
[PubMed:16855244]
[WorldCat.org]
[DOI]
(P p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)