Difference between revisions of "SipW"
| Line 123: | Line 123: | ||
<pubmed>18047568,15175311,16430695,10368135,9694797,18430133,16430695,10464223,17720793, </pubmed> | <pubmed>18047568,15175311,16430695,10368135,9694797,18430133,16430695,10464223,17720793, </pubmed> | ||
| − | |||
Revision as of 14:23, 15 June 2009
- Description: signal peptidase I
| Gene name | sipW |
| Synonyms | yqhE |
| Essential | no |
| Product | signal peptidase I |
| Function | biofilm formation |
| MW, pI | 20 kDa, 5.494 |
| Gene length, protein length | 570 bp, 190 aa |
| Immediate neighbours | tasA, yqxM |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 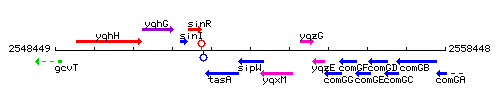
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU24630
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Cleavage of hydrophobic, N-terminal signal or leader sequences from secreted and periplasmic proteins (according to Swiss-Prot)
- Protein family: peptidase S26B family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: secreted (according to Swiss-Prot)
Database entries
- Structure:
- Swiss prot entry: P54506
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References