Difference between revisions of "RecO"
| Line 1: | Line 1: | ||
| − | * '''Description:''' mediator of [[RecA]] binding to ssDNA, required for the formation of [[RecA]] DNA repair centers <br/><br/> | + | * '''Description:''' mediator of [[RecA]] binding to ssDNA, required for the formation of [[RecA]] DNA repair centers, , required for efficient survival and replication restart after replication-transcription conflicts <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 149: | Line 149: | ||
<pubmed> 22933559 </pubmed> | <pubmed> 22933559 </pubmed> | ||
== Original publications == | == Original publications == | ||
| − | <pubmed>24891441,18599486, 19730681 20581116 22373918 17581636 21170359 24285298 24285298</pubmed> | + | <pubmed>24891441,18599486, 19730681 20581116 22373918 17581636 21170359 24285298 24285298 25939832</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 09:15, 7 May 2015
- Description: mediator of RecA binding to ssDNA, required for the formation of RecA DNA repair centers, , required for efficient survival and replication restart after replication-transcription conflicts
| Gene name | recO |
| Synonyms | yqxN, yqfI |
| Essential | no |
| Product | mediator of RecA binding to ssDNA |
| Function | DNA repair/ recombination |
| Gene expression levels in SubtiExpress: recO | |
| Interactions involving this protein in SubtInteract: RecO | |
| MW, pI | 29 kDa, 8.208 |
| Gene length, protein length | 765 bp, 255 aa |
| Immediate neighbours | glyQ, yqzL |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 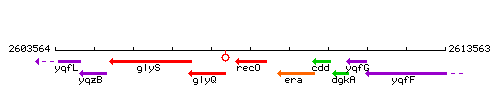
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU25280
Phenotypes of a mutant
- drastically reduced survival of mature dormant spores after exposure to ultrahigh vacuum desiccation and ionizing radiation that induce single strand (ss) DNA nicks and double-strand breaks (DSBs) PubMed
Database entries
- BsubCyc: BSU25280
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- provides RecA access to ssDNA during chromosomal transformation (together with DprA) PubMed
- catalyzes annealing of SsbA or SsbA/SsbB coated ssDNAs to allow the formation of DNA duplexes with tails during plasmid transformation PubMed
- required for the formation of RecA DNA repair centers (together with RecR) PubMed
- Protein family: recO family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cytoplasm (according to Swiss-Prot)
- localizes to the DNA entry pole during transformation PubMed
Database entries
- BsubCyc: BSU25280
- Structure:
- UniProt: P42095
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications