Difference between revisions of "SpsK"
(→Expression and regulation) |
|||
| Line 104: | Line 104: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' ''[[spsA]]-[[spsB]]-[[spsC]]-[[spsD]]-[[spsF]]-[[spsG]]-[[spsI]]-[[spsJ]]-[[spsK]]-[[spsL]]'' {{PubMed|9353933}} | + | * '''Operon:''' ''[[spsA]]-[[spsB]]-[[spsC]]-[[spsD]]-[[spsE]]-[[spsF]]-[[spsG]]-[[spsI]]-[[spsJ]]-[[spsK]]-[[spsL]]'' {{PubMed|9353933}} |
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=spsK_3883427_3884278_-1 spsK] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=spsK_3883427_3884278_-1 spsK] {{PubMed|22383849}} | ||
Revision as of 14:12, 6 April 2015
- Description: dTDP -4-dehydrorhamnose reductase, spore coat polysaccharide synthesis
| Gene name | spsK |
| Synonyms | ipa-73d |
| Essential | no |
| Product | dTDP -4-dehydrorhamnose reductase |
| Function | rhamnose biosynthesis, spore coat polysaccharide synthesis |
| Gene expression levels in SubtiExpress: spsK | |
| MW, pI | 31 kDa, 6.081 |
| Gene length, protein length | 849 bp, 283 aa |
| Immediate neighbours | spsL, spsJ |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 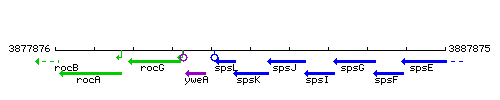
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed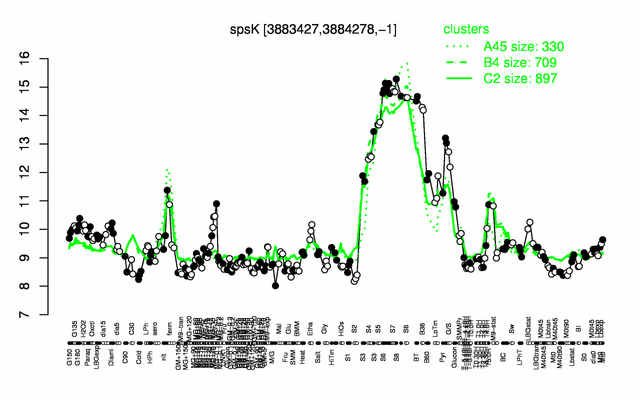
| |
Contents
Categories containing this gene/protein
miscellaneous metabolic pathways, sporulation proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU37820
Phenotypes of a mutant
Database entries
- BsubCyc: BSU37820
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: dTDP-4-dehydrorhamnose reductase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU37820
- Structure: 3SC6 (dTDP-4-dehydrorhamnose reductase (rfbD) from Bacillus anthracis str. Ames in complex with NADP; 42% identity, 72% similarity)
- UniProt: P39631
- KEGG entry: [3]
- E.C. number: 1.1.1.133
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References