Difference between revisions of "RpoC"
(→Extended information on the protein) |
|||
| Line 95: | Line 95: | ||
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
** [[RpoA]]-[[RpoB]]-[[RpoC]] {{PubMed|10499798}} | ** [[RpoA]]-[[RpoB]]-[[RpoC]] {{PubMed|10499798}} | ||
| + | ** [[RpoC]]-[[RpoY]] {{PubMed|25092033}} | ||
** [[SigA]]-([[RpoB]]-[[RpoC]]) {{PubMed|19680289,19735077}}, [[SigB]]-([[RpoB]]-[[RpoC]]) | ** [[SigA]]-([[RpoB]]-[[RpoC]]) {{PubMed|19680289,19735077}}, [[SigB]]-([[RpoB]]-[[RpoC]]) | ||
** [[SigD]]-([[RpoB]]-[[RpoC]]), [[SigE]]-([[RpoB]]-[[RpoC]]) | ** [[SigD]]-([[RpoB]]-[[RpoC]]), [[SigE]]-([[RpoB]]-[[RpoC]]) | ||
Revision as of 11:54, 6 August 2014
- Description: RNA polymerase beta' subunit
| Gene name | rpoC |
| Synonyms | |
| Essential | yes PubMed |
| Product | RNA polymerase beta' subunit |
| Function | transcription |
| Gene expression levels in SubtiExpress: rpoC | |
| Interactions involving this protein in SubtInteract: RpoC | |
| MW, pI | 133 kDa, 8.863 |
| Gene length, protein length | 3597 bp, 1199 aa |
| Immediate neighbours | rpoB, ybxF |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed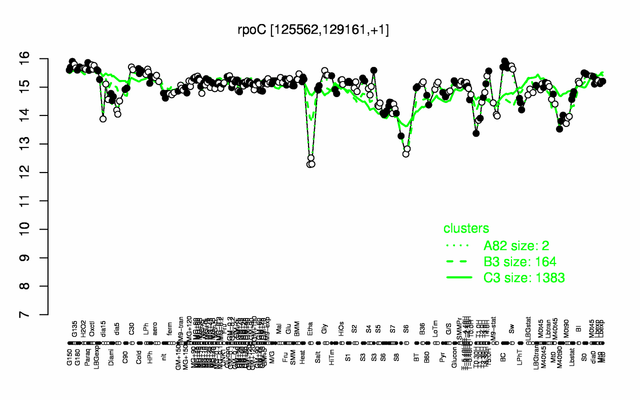
| |
Contents
Categories containing this gene/protein
transcription, essential genes, membrane proteins, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU01080
Phenotypes of a mutant
essential PubMed
Database entries
- BsubCyc: BSU01080
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
- mutations in mtrB, sigB, rpoB, and rpoC allow B. subtilis to grow with 4-fluorotryptophan rather than with tryptophan as a canonical amino acid of the genetic code PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Nucleoside triphosphate + RNA(n) = diphosphate + RNA(n+1) (according to Swiss-Prot)
- Protein family: RNA polymerase beta' chain family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- phosphorylated on Arg-335 and Arg-803 PubMed
- Effectors of protein activity:
- Interactions:
- RpoA-RpoB-RpoC PubMed
- RpoC-RpoY PubMed
- SigA-(RpoB-RpoC) PubMed, SigB-(RpoB-RpoC)
- SigD-(RpoB-RpoC), SigE-(RpoB-RpoC)
- SigF-(RpoB-RpoC), SigG-(RpoB-RpoC)
- SigH-(RpoB-RpoC), SigI-(RpoB-RpoC)
- SigK-(RpoB-RpoC), SigL-(RpoB-RpoC)
- SigM-(RpoB-RpoC), SigV-(RpoB-RpoC)
- SigW-(RpoB-RpoC), SigX-(RpoB-RpoC)
- SigY-(RpoB-RpoC), SigZ-(RpoB-RpoC)
- Xpf-(RpoB-RpoC), YlaC-(RpoB-RpoC)
- YvrHa-RpoC PubMed
Database entries
- BsubCyc: BSU01080
- Structure:
- UniProt: P37871
- KEGG entry: [2]
- E.C. number: 2.7.7.6
Additional information
Expression and regulation
- Operon:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications