Difference between revisions of "GerD"
| Line 57: | Line 57: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU01550&redirect=T BSU01550] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/gerD.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/gerD.html] | ||
| Line 97: | Line 98: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU01550&redirect=T BSU01550] | ||
* '''Structure:''' [http://pdb.org/pdb/explore/explore.do?structureId=4O8W 4O8W] {{PubMed|24530795}} | * '''Structure:''' [http://pdb.org/pdb/explore/explore.do?structureId=4O8W 4O8W] {{PubMed|24530795}} | ||
Revision as of 12:51, 2 April 2014
- Description: scaffold of the germinosome, required for clustering of germinant receptors in the spore inner membrane
| Gene name | gerD |
| Synonyms | |
| Essential | no |
| Product | lipoprotein |
| Function | germination |
| Gene expression levels in SubtiExpress: gerD | |
| Interactions involving this protein in SubtInteract: GerD | |
| MW, pI | 20 kDa, 4.862 |
| Gene length, protein length | 555 bp, 185 aa |
| Immediate neighbours | salA, kbaA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 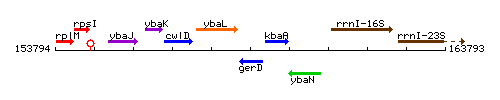
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed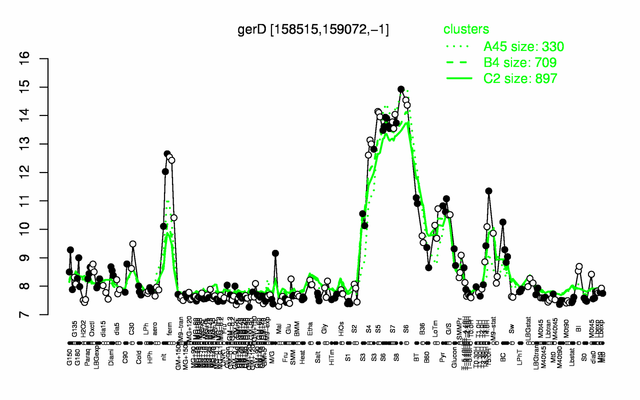
| |
Contents
Categories containing this gene/protein
germination, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU01550
Phenotypes of a mutant
- defective in germination in response to nutrients PubMed
Database entries
- BsubCyc: BSU01550
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: required for clustering of germinant receptors in the spore inner membrane PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- BsubCyc: BSU01550
- UniProt: P16450
- KEGG entry: [3]
- E.C. number:
Additional information
- 3,500 molecules are present per spore PubMed
Expression and regulation
- Regulation:
- expression level depends on sporulation conditions (medim composition, temperature) PubMed
- Regulatory mechanism:
- Additional information:
- 3,500 molecules are present per spore PubMed
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody: available in Anne Moir lab
Labs working on this gene/protein
Anne Moir, University of Sheffield, UK
Your additional remarks
References
Yunfeng Li, Kai Jin, Sonali Ghosh, Parvathimadhavi Devarakonda, Kristina Carlson, Andrew Davis, Kerry-Ann V Stewart, Elizabeth Cammett, Patricia Pelczar Rossi, Barbara Setlow, Min Lu, Peter Setlow, Bing Hao
Structural and functional analysis of the GerD spore germination protein of Bacillus species.
J Mol Biol: 2014, 426(9);1995-2008
[PubMed:24530795]
[WorldCat.org]
[DOI]
(I p)
Kerry-Ann V Stewart, Peter Setlow
Numbers of individual nutrient germinant receptors and other germination proteins in spores of Bacillus subtilis.
J Bacteriol: 2013, 195(16);3575-82
[PubMed:23749970]
[WorldCat.org]
[DOI]
(I p)
George Korza, Peter Setlow
Topology and accessibility of germination proteins in the Bacillus subtilis spore inner membrane.
J Bacteriol: 2013, 195(7);1484-91
[PubMed:23335419]
[WorldCat.org]
[DOI]
(I p)
Kerry-Ann V Stewart, Xuan Yi, Sonali Ghosh, Peter Setlow
Germination protein levels and rates of germination of spores of Bacillus subtilis with overexpressed or deleted genes encoding germination proteins.
J Bacteriol: 2012, 194(12);3156-64
[PubMed:22493018]
[WorldCat.org]
[DOI]
(I p)
Arturo Ramirez-Peralta, Pengfei Zhang, Yong-Qing Li, Peter Setlow
Effects of sporulation conditions on the germination and germination protein levels of Bacillus subtilis spores.
Appl Environ Microbiol: 2012, 78(8);2689-97
[PubMed:22327596]
[WorldCat.org]
[DOI]
(I p)
Xuan Yi, Jintao Liu, James R Faeder, Peter Setlow
Synergism between different germinant receptors in the germination of Bacillus subtilis spores.
J Bacteriol: 2011, 193(18);4664-71
[PubMed:21725007]
[WorldCat.org]
[DOI]
(I p)
Keren K Griffiths, Jingqiao Zhang, Ann E Cowan, Ji Yu, Peter Setlow
Germination proteins in the inner membrane of dormant Bacillus subtilis spores colocalize in a discrete cluster.
Mol Microbiol: 2011, 81(4);1061-77
[PubMed:21696470]
[WorldCat.org]
[DOI]
(I p)
Guiwen Wang, Xuan Yi, Yong-qing Li, Peter Setlow
Germination of individual Bacillus subtilis spores with alterations in the GerD and SpoVA proteins, which are important in spore germination.
J Bacteriol: 2011, 193(9);2301-11
[PubMed:21398556]
[WorldCat.org]
[DOI]
(I p)
Wiyada Mongkolthanaruk, Carl Robinson, Anne Moir
Localization of the GerD spore germination protein in the Bacillus subtilis spore.
Microbiology (Reading): 2009, 155(Pt 4);1146-1151
[PubMed:19332816]
[WorldCat.org]
[DOI]
(P p)
Patricia L Pelczar, Peter Setlow
Localization of the germination protein GerD to the inner membrane in Bacillus subtilis spores.
J Bacteriol: 2008, 190(16);5635-41
[PubMed:18556788]
[WorldCat.org]
[DOI]
(I p)
Patricia L Pelczar, Takao Igarashi, Barbara Setlow, Peter Setlow
Role of GerD in germination of Bacillus subtilis spores.
J Bacteriol: 2007, 189(3);1090-8
[PubMed:17122337]
[WorldCat.org]
[DOI]
(P p)
E H Kemp, R L Sammons, A Moir, D Sun, P Setlow
Analysis of transcriptional control of the gerD spore germination gene of Bacillus subtilis 168.
J Bacteriol: 1991, 173(15);4646-52
[PubMed:1906867]
[WorldCat.org]
[DOI]
(P p)
J R Yon, R L Sammons, D A Smith
Cloning and sequencing of the gerD gene of Bacillus subtilis.
J Gen Microbiol: 1989, 135(12);3431-45
[PubMed:2517635]
[WorldCat.org]
[DOI]
(P p)