Difference between revisions of "SinR"
| Line 74: | Line 74: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' transcription | + | * '''Catalyzed reaction/ biological activity:''' |
| + | ** transcription regulator of biofilm genes, acts as a true repressor of the ''[[tapA]]-[[sipW]]-[[tasA]]'' operon and as an anti-activator (prevents binding of the activator protein [[RemA]]) of the ''[[epsA]]-[[epsB]]-[[epsC]]-[[epsD]]-[[epsE]]-[[epsF]]-[[epsG]]-[[epsH]]-[[epsI]]-[[epsJ]]-[[epsK]]-[[epsL]]-[[epsM]]-[[epsN]]-[[epsO]]'' operon {{PubMed|23646920}} | ||
| + | ** acts as co-repressor for [[SlrR]] {{PubMed|20351052}} | ||
* '''Protein family:''' | * '''Protein family:''' | ||
Revision as of 15:07, 24 May 2013
- Description: transcriptional regulator of post-exponential-phase responses genes
| Gene name | sinR |
| Synonyms | sin, flaD |
| Essential | no |
| Product | transcriptional regulator of post-exponential-phase responses genes |
| Function | control of biofilm formation |
| Gene expression levels in SubtiExpress: sinR | |
| Interactions involving this protein in SubtInteract: SinR | |
| Metabolic function and regulation of this protein in SubtiPathways: Biofilm, Central C-metabolism, Protein secretion | |
| MW, pI | 12 kDa, 7.177 |
| Gene length, protein length | 333 bp, 111 aa |
| Immediate neighbours | sinI, tasA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 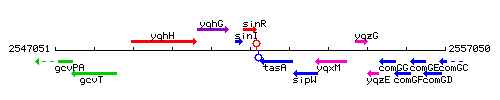
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
transcription factors and their control, transition state regulators, biofilm formation
This gene is a member of the following regulons
AbrB regulon, ScoC regulon, Spo0A regulon
The SinR regulon
The gene
Basic information
- Locus tag: BSU24610
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- transcription regulator of biofilm genes, acts as a true repressor of the tapA-sipW-tasA operon and as an anti-activator (prevents binding of the activator protein RemA) of the epsA-epsB-epsC-epsD-epsE-epsF-epsG-epsH-epsI-epsJ-epsK-epsL-epsM-epsN-epsO operon PubMed
- acts as co-repressor for SlrR PubMed
- Protein family:
- Paralogous protein(s): SlrR
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P06533
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: TMB079 sinR::spec, GP736 (tetR), available in Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Modelling of the SinI/SinR switch
Original publications