Difference between revisions of "CdaA"
| Line 4: | Line 4: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''cdaA'' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''ybbQ '' | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''ybbP, ybbQ '' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
| Line 14: | Line 14: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || synthesis of c-di-AMP | |style="background:#ABCDEF;" align="center"|'''Function''' || synthesis of c-di-AMP | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU01750 | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU01750 cdaA] |
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/CdaA CdaA] | ||
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/murein/index.html Murein recycling]''' | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/murein/index.html Murein recycling]''' | ||
| Line 22: | Line 24: | ||
|style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 819 bp, 273 aa | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 819 bp, 273 aa | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ybbM]]'', ''[[ | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ybbM]]'', ''[[cdaR]]'' |
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS:CAB11951]+-newId sequences] <br/> (Barbe ''et al.'', 2009)''' | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS:CAB11951]+-newId sequences] <br/> (Barbe ''et al.'', 2009)''' | ||
| Line 55: | Line 57: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| − | * inactivation of ''[[ | + | * inactivation of ''[[cdaA]]'' results in severe beta-lactam sensitivity {{PubMed|22211522}} |
=== Database entries === | === Database entries === | ||
| Line 73: | Line 75: | ||
* '''Protein family:''' | * '''Protein family:''' | ||
| − | * '''Paralogous protein(s):''' [[ | + | * '''Paralogous protein(s):''' [[CdaS]] |
=== Extended information on the protein === | === Extended information on the protein === | ||
| Line 86: | Line 88: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| + | ** the interaction with [[CdaR]] stimulates the diadenylate cyclase activity of [[CdaA]] {{PubMed|23192352}} | ||
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
| + | ** [[CdaA]]-[[CdaR]], to stimulate [[CdaA]] activity {{PubMed|23192352}} | ||
* '''[[Localization]]:''' | * '''[[Localization]]:''' | ||
| Line 108: | Line 112: | ||
* '''Operon:''' ''[[cdaA]]-[[cdaR]]-[[glmM]]-[[glmS]]'' {{PubMed|23192352}} | * '''Operon:''' ''[[cdaA]]-[[cdaR]]-[[glmM]]-[[glmS]]'' {{PubMed|23192352}} | ||
| − | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=ybbP_196213_197034_1 | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=ybbP_196213_197034_1 cdaA] {{PubMed|22383849}} |
* '''[[Sigma factor]]:''' [[SigA]] {{PubMed|22211522}} | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|22211522}} | ||
| Line 116: | Line 120: | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | * '''Additional information:''' the ''[[ | + | * '''Additional information:''' the ''[[cdaA]]-[[cdaR]]'' part of the mRNA is very stable (half-life > 15 min) [http://www.ncbi.nlm.nih.gov/sites/entrez/12884008 PubMed] |
=Biological materials = | =Biological materials = | ||
* '''Mutant:''' | * '''Mutant:''' | ||
| − | ** GP997 (''[[ | + | ** GP997 (''[[cdaA]]''::''cat''), available in [[Jörg Stülke]]'s lab |
| − | ** GP985 (''[[ | + | ** GP985 (''[[cdaA]]''-''[[cdaR]]''::''cat''), available in [[Jörg Stülke]]'s lab |
* '''Expression vector:''' | * '''Expression vector:''' | ||
| − | ** expression of native '' | + | ** expression of native ''cdaA'' in ''B. subtilis'': pGP1960 (in [[pBQ200]]), available in [[Jörg Stülke]]'s lab |
| − | ** expression of '' | + | ** expression of ''cdaA''-Strep in ''B. subtilis'' suitable for [[SPINE]]: pGP1986 (in [[pGP382]]), available in [[Jörg Stülke]]'s lab |
* '''lacZ fusion:''' GP1339 (cat) based on [[pAC6]], available in [[Jörg Stülke]]'s lab | * '''lacZ fusion:''' GP1339 (cat) based on [[pAC6]], available in [[Jörg Stülke]]'s lab | ||
| Line 135: | Line 139: | ||
* '''FLAG-tag construct:''' | * '''FLAG-tag construct:''' | ||
| − | ** GP1331 (''[[ | + | ** GP1331 (''[[cdaA]]''-3xFLAG ''ermC'', based on [[pGP1087]]), available in [[Jörg Stülke]]'s lab |
* '''Antibody:''' | * '''Antibody:''' | ||
Revision as of 11:45, 30 November 2012
- Description: diadenylate cyclase, synthesis of c-di-AMP in vegetative cells
| Gene name | cdaA |
| Synonyms | ybbP, ybbQ |
| Essential | no |
| Product | diadenylate cyclase |
| Function | synthesis of c-di-AMP |
| Gene expression levels in SubtiExpress: cdaA | |
| Interactions involving this protein in SubtInteract: CdaA | |
| Metabolic function and regulation of this protein in SubtiPathways: Murein recycling | |
| MW, pI | 30 kDa, 8.074 |
| Gene length, protein length | 819 bp, 273 aa |
| Immediate neighbours | ybbM, cdaR |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 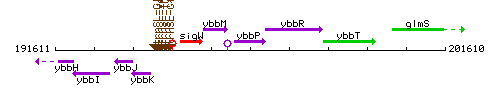
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
membrane proteins, metabolism of signalling nucleotides
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU01750
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s): CdaS
Extended information on the protein
- Kinetic information:
- Domains: contains a DAC domain potentially involved in the synthesis of c-di-AMP PubMed
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: Q45589
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
Biological materials
- Mutant:
- GP997 (cdaA::cat), available in Jörg Stülke's lab
- GP985 (cdaA-cdaR::cat), available in Jörg Stülke's lab
- Expression vector:
- expression of native cdaA in B. subtilis: pGP1960 (in pBQ200), available in Jörg Stülke's lab
- expression of cdaA-Strep in B. subtilis suitable for SPINE: pGP1986 (in pGP382), available in Jörg Stülke's lab
- lacZ fusion: GP1339 (cat) based on pAC6, available in Jörg Stülke's lab
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's lab. Respective plasmid: pGP1990.
- FLAG-tag construct:
- GP1331 (cdaA-3xFLAG ermC, based on pGP1087), available in Jörg Stülke's lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications
Additional publications: PubMed