Difference between revisions of "ClpC"
m (Reverted edits by 134.76.70.252 (talk) to last revision by Jstuelk) |
|||
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || protein degradation<br/>positive regulator of autolysin ([[LytC]] and [[LytD]]) synthesis | |style="background:#ABCDEF;" align="center"|'''Function''' || protein degradation<br/>positive regulator of autolysin ([[LytC]] and [[LytD]]) synthesis | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU00860 clpC] | ||
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/ClpC ClpC] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/ClpC ClpC] | ||
Revision as of 08:56, 8 August 2012
- Description: ATPase subunit of the ATP-dependent ClpC-ClpP protease, involved in competence development, heat shock regulation, motility, sporulation, protein quality control, biofilm formation
| Gene name | clpC |
| Synonyms | mecB |
| Essential | no |
| Product | ATPase subunit of the ClpC-ClpP protease |
| Function | protein degradation positive regulator of autolysin (LytC and LytD) synthesis |
| Gene expression levels in SubtiExpress: clpC | |
| Interactions involving this protein in SubtInteract: ClpC | |
| Metabolic function and regulation of this protein in SubtiPathways: Stress | |
| MW, pI | 89 kDa, 5.746 |
| Gene length, protein length | 2430 bp, 810 aa |
| Immediate neighbours | mcsB, radA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 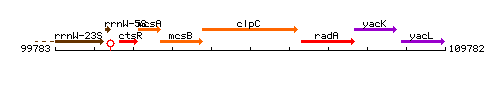
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed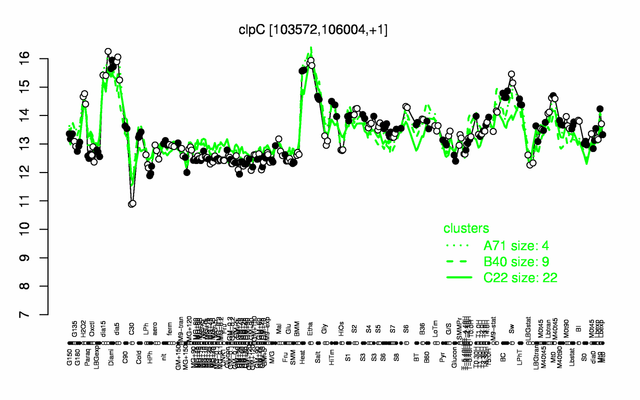
| |
Contents
Categories containing this gene/protein
proteolysis, sporulation proteins, general stress proteins (controlled by SigB), heat shock proteins, phosphoproteins
This gene is a member of the following regulons
CtsR regulon, SigB regulon, SigF regulon
The gene
Basic information
- Locus tag: BSU00860
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- A mutation was found in this gene after evolution under relaxed selection for sporulation PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATPase/chaperone
- Protein family: mecA family (according to Swiss-Prot) clpA/clpB family. ClpC subfamily (according to Swiss-Prot), AAA+ -type ATPase (IPR013093) InterPro (PF07724) PFAM
Targets of ClpC-ClpP-dependent protein degradation
Extended information on the protein
- Kinetic information:
- Domains: AAA-ATPase PFAM
- Modification:
- phosphorylated on Arg-5 and Arg-254 PubMed
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: P37571
- KEGG entry: [3]
- E.C. number:
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Regulation:
- Additional information: subject to Clp-dependent proteolysis upon glucose starvation PubMed
Biological materials
- Expression vector:
- lacZ fusion:
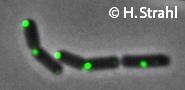
- GFP fusion: C-terminal GFP fusions (single copy, also as CFP and YFP variants) available from the Hamoen Lab
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Leendert Hamoen, Newcastle University, UK homepage
Kürsad Turgay, Freie Universität Berlin, Germany homepage
Your additional remarks
References
Reviews
Additional reviews: PubMed
Dorte Frees, Kirsi Savijoki, Pekka Varmanen, Hanne Ingmer
Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria.
Mol Microbiol: 2007, 63(5);1285-95
[PubMed:17302811]
[WorldCat.org]
[DOI]
(P p)
Original Publications
Additional publications: PubMed