Difference between revisions of "FruA"
m (Reverted edits by 134.76.70.252 (talk) to last revision by Jstuelk) |
|||
| Line 104: | Line 104: | ||
* '''Operon:''' ''[[fruR]]-[[fruK]]-[[fruA]]'' (according to [http://dbtbs.hgc.jp/COG/prom/fruRKA.html DBTBS]) | * '''Operon:''' ''[[fruR]]-[[fruK]]-[[fruA]]'' (according to [http://dbtbs.hgc.jp/COG/prom/fruRKA.html DBTBS]) | ||
| − | * '''[ | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=fruA_1509256_1511163_1 fruA] {{PubMed|22383849}} |
| + | |||
| + | * '''Sigma factor:''' | ||
* '''Regulation:''' | * '''Regulation:''' | ||
Revision as of 08:04, 13 April 2012
- Description: fructose-specific phosphotransferase system, EIIABC of the PTS
| Gene name | fruA |
| Synonyms | |
| Essential | no |
| Product | fructose-specific phosphotransferase system, EIIABC |
| Function | fructose uptake and phosphorylation |
| Interactions involving this protein in SubtInteract: FruA | |
| Metabolic function and regulation of this protein in SubtiPathways: Sugar catabolism | |
| MW, pI | 67 kDa, 5.241 |
| Gene length, protein length | 1905 bp, 635 aa |
| Immediate neighbours | fruK, sipT |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 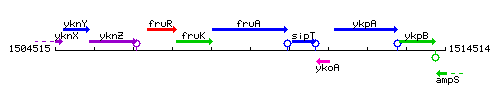
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
phosphotransferase systems, utilization of specific carbon sources, membrane proteins, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU14400
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Protein EIIA N(pi)-phospho-L-histidine + protein EIIB = protein EIIA + protein EIIB N(pi)-phospho-L-histidine/cysteine (according to Swiss-Prot)
- Paralogous protein(s): ManP
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- FruA-HPr
- FruA is a member of a suspected group of hubs proteins that were suggested to be involved in a large number of interactions PubMed
- Localization: cell membrane (homogeneous) PubMed
Database entries
- Structure: 2R4Q
- UniProt: P71012
- KEGG entry: [3]
- E.C. number: 2.7.1.69
Additional information
Expression and regulation
- Sigma factor:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Elodie Marchadier, Rut Carballido-López, Sophie Brinster, Céline Fabret, Peggy Mervelet, Philippe Bessières, Marie-Françoise Noirot-Gros, Vincent Fromion, Philippe Noirot
An expanded protein-protein interaction network in Bacillus subtilis reveals a group of hubs: Exploration by an integrative approach.
Proteomics: 2011, 11(15);2981-91
[PubMed:21630458]
[WorldCat.org]
[DOI]
(I p)
Jean-Christophe Meile, Ling Juan Wu, S Dusko Ehrlich, Jeff Errington, Philippe Noirot
Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory.
Proteomics: 2006, 6(7);2135-46
[PubMed:16479537]
[WorldCat.org]
[DOI]
(P p)
Jonathan Reizer, Steffi Bachem, Aiala Reizer, Maryvonne Arnaud, Milton H Saier, Jörg Stülke
Novel phosphotransferase system genes revealed by genome analysis - the complete complement of PTS proteins encoded within the genome of Bacillus subtilis.
Microbiology (Reading): 1999, 145 ( Pt 12);3419-3429
[PubMed:10627040]
[WorldCat.org]
[DOI]
(P p)