Difference between revisions of "YvmC"
| Line 53: | Line 53: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
| Line 86: | Line 83: | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' | + | * '''Structure:''' [http://www.pdb.org/pdb/explore/explore.do?structureId=3S7T 3S7T] (from ''B. licheniformis'', 70% identity, 86% similarity) {{PubMed|21325056}} |
* '''UniProt:''' [http://www.uniprot.org/uniprot/O34351 O34351] | * '''UniProt:''' [http://www.uniprot.org/uniprot/O34351 O34351] | ||
| Line 127: | Line 124: | ||
=References= | =References= | ||
| − | <pubmed> 19430487 4204912 20817675</pubmed> | + | <pubmed> 19430487 4204912 20817675 21325056</pubmed> |
[http://www.liebertonline.com/doi/pdf/10.1089/ind.2006.2.66 additional paper] | [http://www.liebertonline.com/doi/pdf/10.1089/ind.2006.2.66 additional paper] | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 20:37, 29 June 2011
- Description: cyclodipeptide synthase
| Gene name | yvmC |
| Synonyms | |
| Essential | no |
| Product | cyclodipeptide synthase |
| Function | biosynthesis of the extracellular iron chelate pulcherrimin |
| MW, pI | 28 kDa, 6.612 |
| Gene length, protein length | 744 bp, 248 aa |
| Immediate neighbours | cypX, yvmB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 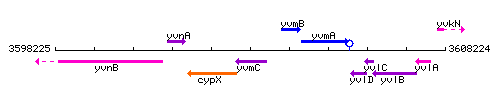
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
acquisition of iron, iron metabolism
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU35070
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- synthesis of the cyclic dipeptide cyclo-L-leucyl-L-leucyl with the corresponding charged tRNAs as substrates in an ATP-dependent manner PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- UniProt: O34351
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Luc Bonnefond, Taiga Arai, Yuriko Sakaguchi, Tsutomu Suzuki, Ryuichiro Ishitani, Osamu Nureki
Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog.
Proc Natl Acad Sci U S A: 2011, 108(10);3912-7
[PubMed:21325056]
[WorldCat.org]
[DOI]
(I p)
Onuma Chumsakul, Hiroki Takahashi, Taku Oshima, Takahiro Hishimoto, Shigehiko Kanaya, Naotake Ogasawara, Shu Ishikawa
Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation.
Nucleic Acids Res: 2011, 39(2);414-28
[PubMed:20817675]
[WorldCat.org]
[DOI]
(I p)
Muriel Gondry, Ludovic Sauguet, Pascal Belin, Robert Thai, Rachel Amouroux, Carine Tellier, Karine Tuphile, Mickaël Jacquet, Sandrine Braud, Marie Courçon, Cédric Masson, Steven Dubois, Sylvie Lautru, Alain Lecoq, Shin-ichi Hashimoto, Roger Genet, Jean-Luc Pernodet
Cyclodipeptide synthases are a family of tRNA-dependent peptide bond-forming enzymes.
Nat Chem Biol: 2009, 5(6);414-20
[PubMed:19430487]
[WorldCat.org]
[DOI]
(I p)
R L Uffen, E Canale-Parola
Synthesis of pulcherriminic acid by Bacillus subtilis.
J Bacteriol: 1972, 111(1);86-93
[PubMed:4204912]
[WorldCat.org]
[DOI]
(P p)