Difference between revisions of "SpoIIAA"
(→Expression and regulation) |
|||
| Line 90: | Line 90: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' ''[[dacF]]-[[spoIIAA]]-[[spoIIAB]]-[[sigF]]'' | + | * '''Operon:''' |
| − | ''[[spoIIAA]]-[[spoIIAB]]-[[sigF]]'' | + | ** ''[[dacF]]-[[spoIIAA]]-[[spoIIAB]]-[[sigF]]'' |
| + | ** ''[[spoIIAA]]-[[spoIIAB]]-[[sigF]]'' | ||
| − | * '''[[Sigma factor]]:''' ''[[dacF]]'': [[SigF]], [[SigG]] | + | * '''[[Sigma factor]]:''' |
| − | ''[[spoIIAA]]'': [[SigH]] | + | ** ''[[dacF]]'': [[SigF]], [[SigG]] |
| + | ** ''[[spoIIAA]]'': [[SigH]] | ||
* '''Regulation:''' repressed by glucose (5.2-fold) [http://www.ncbi.nlm.nih.gov/pubmed/12850135 PubMed], ''[[dacF]]'': expressed during sporulation, ''[[spoIIAA]]'': expressed early during sporulation | * '''Regulation:''' repressed by glucose (5.2-fold) [http://www.ncbi.nlm.nih.gov/pubmed/12850135 PubMed], ''[[dacF]]'': expressed during sporulation, ''[[spoIIAA]]'': expressed early during sporulation | ||
| Line 127: | Line 129: | ||
=References= | =References= | ||
| − | <pubmed>1391042,8358793,8955289,15023063,15351644,9077448,10323866,8820658,8622920,16824103,7570023,10476035,8932322,8764398,8764397,12676949,9826498,9826499,14744853,9642177,11684022,8846916,11298272,1948031,3114419,11846550,15201047,2123551,8168129,,1556084,15687200,12850135 16493705 17218307, </pubmed> | + | <pubmed>1391042,8358793,8955289,15023063,15351644,9077448,10323866,8820658,8622920,16824103,7570023,10476035,8932322,8764398,8764397,12676949,9826498,9826499,14744853,9642177,11684022,8846916,11298272,1948031,3114419,11846550,15201047,2123551,8168129,,1556084,15687200,12850135 16493705 17218307, 20298743 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 08:30, 20 March 2010
- Description: anti-anti-SigF
| Gene name | spoIIAA |
| Synonyms | |
| Essential | no |
| Product | anti-anti-SigF |
| Function | control of SigF activity |
| Metabolic function and regulation of this protein in SubtiPathways: Stress | |
| MW, pI | 12 kDa, 5.855 |
| Gene length, protein length | 351 bp, 117 aa |
| Immediate neighbours | spoIIAB, dacF |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 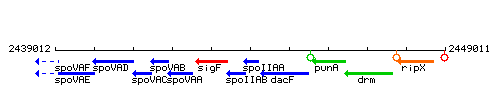
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU23470
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: anti-sigma-factor antagonist family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- UniProt: P10727
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation: repressed by glucose (5.2-fold) PubMed, dacF: expressed during sporulation, spoIIAA: expressed early during sporulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Tony Wilkinson, York University, U.K. homepage
- Charles Moran, Emory University, NC, USA homepage
Your additional remarks
References
Jin Hwan Do, Masao Nagasaki, Satoru Miyano
The systems approach to the prespore-specific activation of sigma factor SigF in Bacillus subtilis.
Biosystems: 2010, 100(3);178-84
[PubMed:20298743]
[WorldCat.org]
[DOI]
(I p)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Oleg A Igoshin, Chester W Price, Michael A Savageau
Signalling network with a bistable hysteretic switch controls developmental activation of the sigma transcription factor in Bacillus subtilis.
Mol Microbiol: 2006, 61(1);165-84
[PubMed:16824103]
[WorldCat.org]
[DOI]
(P p)
Alain Lévine, Françoise Vannier, Cédric Absalon, Lauriane Kuhn, Peter Jackson, Elaine Scrivener, Valérie Labas, Joëlle Vinh, Patrick Courtney, Jérôme Garin, Simone J Séror
Analysis of the dynamic Bacillus subtilis Ser/Thr/Tyr phosphoproteome implicated in a wide variety of cellular processes.
Proteomics: 2006, 6(7);2157-73
[PubMed:16493705]
[WorldCat.org]
[DOI]
(P p)
Masaya Fujita, José Eduardo González-Pastor, Richard Losick
High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis.
J Bacteriol: 2005, 187(4);1357-68
[PubMed:15687200]
[WorldCat.org]
[DOI]
(P p)
Joanna Clarkson, Iain D Campbell, Michael D Yudkin
Efficient regulation of sigmaF, the first sporulation-specific sigma factor in B.subtilis.
J Mol Biol: 2004, 342(4);1187-95
[PubMed:15351644]
[WorldCat.org]
[DOI]
(P p)
Joanna Clarkson, Iain D Campbell, Michael D Yudkin
Physical evidence for the induced release of the Bacillus subtilis transcription factor, sigma(F), from its inhibitory complex.
J Mol Biol: 2004, 340(2);203-9
[PubMed:15201047]
[WorldCat.org]
[DOI]
(P p)
Joanna Clarkson, Jwu-Ching Shu, David A Harris, Iain D Campbell, Michael D Yudkin
Fluorescence and kinetic analysis of the SpoIIAB phosphorylation reaction, a key regulator of sporulation in Bacillus subtilis.
Biochemistry: 2004, 43(11);3120-8
[PubMed:15023063]
[WorldCat.org]
[DOI]
(P p)
Karen Carniol, Patrick Eichenberger, Richard Losick
A threshold mechanism governing activation of the developmental regulatory protein sigma F in Bacillus subtilis.
J Biol Chem: 2004, 279(15);14860-70
[PubMed:14744853]
[WorldCat.org]
[DOI]
(P p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
Margaret S Ho, Karen Carniol, Richard Losick
Evidence in support of a docking model for the release of the transcription factor sigma F from the antisigma factor SpoIIAB in Bacillus subtilis.
J Biol Chem: 2003, 278(23);20898-905
[PubMed:12676949]
[WorldCat.org]
[DOI]
(P p)
J Clarkson, I D Campbell, M D Yudkin
NMR studies of the interactions of SpoIIAA with its partner proteins that regulate sporulation in Bacillus subtilis.
J Mol Biol: 2001, 314(3);359-64
[PubMed:11846550]
[WorldCat.org]
[DOI]
(P p)
Q Pan, D A Garsin, R Losick
Self-reinforcing activation of a cell-specific transcription factor by proteolysis of an anti-sigma factor in B. subtilis.
Mol Cell: 2001, 8(4);873-83
[PubMed:11684022]
[WorldCat.org]
[DOI]
(P p)
C S Lee, J Clarkson, J C Shu, I D Campbell, M D Yudkin
Bacillus subtilis mutations that alter the pathway of phosphorylation of the anti-anti-sigmaF factor SpoIIAA lead to a Spo- phenotype.
Mol Microbiol: 2001, 40(1);9-19
[PubMed:11298272]
[WorldCat.org]
[DOI]
(P p)
A Feucht, R A Daniel, J Errington
Characterization of a morphological checkpoint coupling cell-specific transcription to septation in Bacillus subtilis.
Mol Microbiol: 1999, 33(5);1015-26
[PubMed:10476035]
[WorldCat.org]
[DOI]
(P p)
N King, O Dreesen, P Stragier, K Pogliano, R Losick
Septation, dephosphorylation, and the activation of sigmaF during sporulation in Bacillus subtilis.
Genes Dev: 1999, 13(9);1156-67
[PubMed:10323866]
[WorldCat.org]
[DOI]
(P p)
D A Garsin, L Duncan, D M Paskowitz, R Losick
The kinase activity of the antisigma factor SpoIIAB is required for activation as well as inhibition of transcription factor sigmaF during sporulation in Bacillus subtilis.
J Mol Biol: 1998, 284(3);569-78
[PubMed:9826499]
[WorldCat.org]
[DOI]
(P p)
D A Garsin, D M Paskowitz, L Duncan, R Losick
Evidence for common sites of contact between the antisigma factor SpoIIAB and its partners SpoIIAA and the developmental transcription factor sigmaF in Bacillus subtilis.
J Mol Biol: 1998, 284(3);557-68
[PubMed:9826498]
[WorldCat.org]
[DOI]
(P p)
P J Lewis, L J Wu, J Errington
Establishment of prespore-specific gene expression in Bacillus subtilis: localization of SpoIIE phosphatase and initiation of compartment-specific proteolysis.
J Bacteriol: 1998, 180(13);3276-84
[PubMed:9642177]
[WorldCat.org]
[DOI]
(P p)
M Lord, T Magnin, M D Yudkin
Protein conformational change and nucleotide binding involved in regulation of sigmaF in Bacillus subtilis.
J Bacteriol: 1996, 178(23);6730-5
[PubMed:8955289]
[WorldCat.org]
[DOI]
(P p)
S M Najafi, D A Harris, M D Yudkin
The SpoIIAA protein of Bacillus subtilis has GTP-binding properties.
J Bacteriol: 1996, 178(22);6632-4
[PubMed:8932322]
[WorldCat.org]
[DOI]
(P p)
P J Lewis, T Magnin, J Errington
Compartmentalized distribution of the proteins controlling the prespore-specific transcription factor sigmaF of Bacillus subtilis.
Genes Cells: 1996, 1(10);881-94
[PubMed:9077448]
[WorldCat.org]
[DOI]
(P p)
S Alper, A Dufour, D A Garsin, L Duncan, R Losick
Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis.
J Mol Biol: 1996, 260(2);165-77
[PubMed:8764398]
[WorldCat.org]
[DOI]
(P p)
L Duncan, S Alper, R Losick
SpoIIAA governs the release of the cell-type specific transcription factor sigma F from its anti-sigma factor SpoIIAB.
J Mol Biol: 1996, 260(2);147-64
[PubMed:8764397]
[WorldCat.org]
[DOI]
(P p)
F Arigoni, L Duncan, S Alper, R Losick, P Stragier
SpoIIE governs the phosphorylation state of a protein regulating transcription factor sigma F during sporulation in Bacillus subtilis.
Proc Natl Acad Sci U S A: 1996, 93(8);3238-42
[PubMed:8622920]
[WorldCat.org]
[DOI]
(P p)
A Feucht, T Magnin, M D Yudkin, J Errington
Bifunctional protein required for asymmetric cell division and cell-specific transcription in Bacillus subtilis.
Genes Dev: 1996, 10(7);794-803
[PubMed:8846916]
[WorldCat.org]
[DOI]
(P p)
T Magnin, M Lord, J Errington, M D Yudkin
Establishing differential gene expression in sporulating Bacillus subtilis: phosphorylation of SpoIIAA (anti-anti-sigmaF) alters its conformation and prevents formation of a SpoIIAA/SpoIIAB/ADP complex.
Mol Microbiol: 1996, 19(4);901-7
[PubMed:8820658]
[WorldCat.org]
[DOI]
(P p)
L Duncan, S Alper, F Arigoni, R Losick, P Stragier
Activation of cell-specific transcription by a serine phosphatase at the site of asymmetric division.
Science: 1995, 270(5236);641-4
[PubMed:7570023]
[WorldCat.org]
[DOI]
(P p)
S Alper, L Duncan, R Losick
An adenosine nucleotide switch controlling the activity of a cell type-specific transcription factor in B. subtilis.
Cell: 1994, 77(2);195-205
[PubMed:8168129]
[WorldCat.org]
[DOI]
(P p)
K T Min, C M Hilditch, B Diederich, J Errington, M D Yudkin
Sigma F, the first compartment-specific transcription factor of B. subtilis, is regulated by an anti-sigma factor that is also a protein kinase.
Cell: 1993, 74(4);735-42
[PubMed:8358793]
[WorldCat.org]
[DOI]
(P p)
T Bird, D Burbulys, J J Wu, M A Strauch, J A Hoch, G B Spiegelman
The effect of supercoiling on the in vitro transcription of the spoIIA operon from Bacillus subtilis.
Biochimie: 1992, 74(7-8);627-34
[PubMed:1391042]
[WorldCat.org]
[DOI]
(P p)
K York, T J Kenney, S Satola, C P Moran, H Poth, P Youngman
Spo0A controls the sigma A-dependent activation of Bacillus subtilis sporulation-specific transcription unit spoIIE.
J Bacteriol: 1992, 174(8);2648-58
[PubMed:1556084]
[WorldCat.org]
[DOI]
(P p)
P Margolis, A Driks, R Losick
Establishment of cell type by compartmentalized activation of a transcription factor.
Science: 1991, 254(5031);562-5
[PubMed:1948031]
[WorldCat.org]
[DOI]
(P p)
R Schmidt, P Margolis, L Duncan, R Coppolecchia, C P Moran, R Losick
Control of developmental transcription factor sigma F by sporulation regulatory proteins SpoIIAA and SpoIIAB in Bacillus subtilis.
Proc Natl Acad Sci U S A: 1990, 87(23);9221-5
[PubMed:2123551]
[WorldCat.org]
[DOI]
(P p)
J Errington, J Mandelstam
Use of a lacZ gene fusion to determine the dependence pattern of sporulation operon spoIIA in spo mutants of Bacillus subtilis.
J Gen Microbiol: 1986, 132(11);2967-76
[PubMed:3114419]
[WorldCat.org]
[DOI]
(P p)