GudB
- Description: trigger enzyme: glutamate dehydrogenase (cryptic in 168 and derivatives)
| Gene name | gudB |
| Synonyms | ypcA |
| Essential | no |
| Product | trigger enzyme: glutamate dehydrogenase |
| Function | glutamate utilization, control of GltC activity |
| Gene expression levels in SubtiExpress: gudB | |
| Interactions involving this protein in SubtInteract: GudB | |
| Metabolic function and regulation of this protein in SubtiPathways: gudB | |
| MW, pI | 47 kDa, 5.582 |
| Gene length, protein length | 1278 bp, 426 aa |
| Immediate neighbours | ypdA, ypbH |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 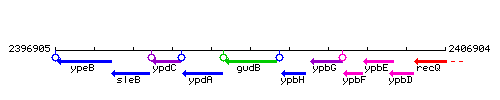
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
utilization of amino acids, glutamate metabolism, transcription factors and their control, trigger enzyme, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU22960
Phenotypes of a mutant
- The gene is cryptic. If gudB is activated (gudB1 mutation), the bacteria are able to utilize glutamate as the only carbon source. PubMed
- A rocG gudB mutant is sensitive to ß-lactam antibiotics such as cefuroxime and to fosfomycin due to the downregulation of the SigW regulon PubMed
- transcription profile of a rocG gudB mutant strain: GEO PubMed
Database entries
- BsubCyc: BSU22960
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: L-glutamate + H2O + NAD+ = 2-oxoglutarate + NH3 + NADH + H+ (according to Swiss-Prot)
- Protein family: Glu/Leu/Phe/Val dehydrogenases family (according to Swiss-Prot)
- Paralogous protein(s): RocG
Extended information on the protein
- Kinetic information:
- Modification:
- phosphorylated on Arg-56, Arg-83, and Arg-421 and/or Arg-423 PubMed
- Cofactors: NAD+/NADH + H+
- Effectors of protein activity:
Database entries
- BsubCyc: BSU22960
- UniProt: P50735
- KEGG entry: [4]
- E.C. number: 1.4.1.2
Additional information
Expression and regulation
- Operon: gudB PubMed
- Regulation: constitutively expressed PubMed
- Regulatory mechanism:
- Additional information: GudB is subject to Clp-dependent proteolysis upon glucose starvation PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 239 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 6565 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 1157 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 580 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 888 PubMed
Biological materials
- Mutant: GP691 (ΔgudB::cat), GP1160 (ΔgudB::aphA3) both available in Jörg Stülke's lab
- Mutant: BP442 (ΔgudB::aphA3), lacking the complete promoter, available in Fabian Commichau's lab
- Expression vector:
- for purification of GudB from E. coli carrying an N-terminal Strep-tag: pGP863 (in pGP172) available in Jörg Stülke's lab
- for purification of GudB1 from E. coli carrying an N-terminal Strep-tag: pGP864 (in pGP172) available in Jörg Stülke's lab
- for ectopic expression of gudB with its native promoter: pGP900 (in pAC5), available in Jörg Stülke's lab
- wild type gudB, expression in B. subtilis, in pBQ200: pGP1712, available in Jörg Stülke's lab
- lacZ fusion: pGP651 (in pAC5), available in Jörg Stülke's lab
- FLAG-tag construct: GP1194 (gudB, spc, based on pGP1331), GP1195 (gudB1, spc, based on pGP1331), available in Jörg Stülke's lab
- GFP fusion:
- two-hybrid system:
- Antibody: antibody against RocG recognizes GudB, available in Jörg Stülke's lab
Labs working on this gene/protein
- Linc Sonenshein, Tufts University, Boston, MA, USA Homepage
- Fabian Commichau University of Göttingen, Germany Homepage
Your additional remarks
The GudB protein is active in other legacy B. subtilis strains (e.g. strain 122). Thus, it can be speculated that the ancestral gudB gene was not cryptic, but became so as a product of the "domestication" of B. subtilis 168 in the lab. PubMed
References
Reviews
Original publications