SsrA
- Description: tmRNA
| Gene name | ssrA |
| Synonyms | |
| Essential | no |
| Product | tmRNA |
| Function | trans-translation |
| Gene length | bp |
| Immediate neighbours | yvaG, smpB |
| Gene sequence (+200bp) | |
Genetic context 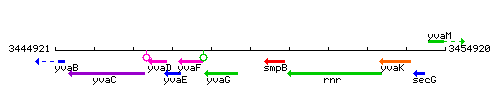
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag:
Phenotypes of a mutant
- impaired growth at high and low temperatures PubMed
- inactivation of ssrA restores beta-lactam resistance in a sigM mutant PubMed
Database entries
- BsubCyc: BSU_misc_RNA_55
- DBTBS entry: no entry
- SubtiList entry: [1]
- Structure:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant: GP323 (cat), available in the Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/RNA
- Chet Price, Davis, USA homepage
Your additional remarks
References
Reviews
Original publications