Sandbox
- Description: DNA gyrase (subunit A)
| Gene name | gyrA |
| Synonyms | nalA |
| Essential | yes PubMed |
| Product | DNA gyrase (subunit A) |
| Function | DNA supercoiling, initation of replication cycle and DNA elongation |
| Gene expression levels in SubtiExpress: GyrA | |
| Interactions involving this protein in SubtInteract: GyrA | |
| MW, pI | 91 kDa, 5.215 |
| Gene length, protein length | 2463 bp, 821 aa |
| Immediate neighbours | gyrB, rrnO-16S |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 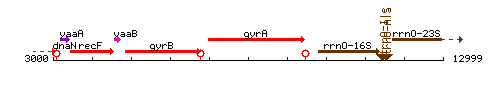
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed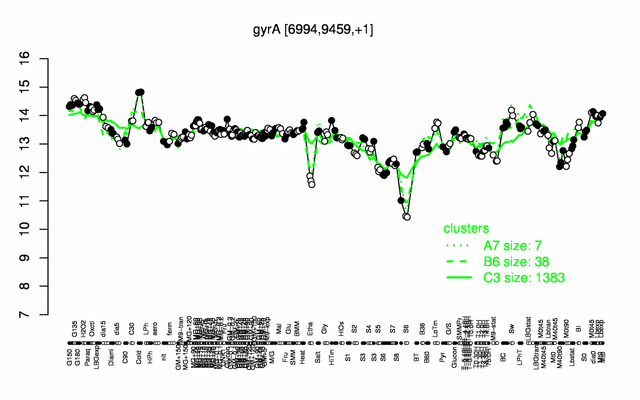
| |
Contents
Categories containing this gene/protein
DNA condensation/ segregation, essential genes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU00070
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP-dependent breakage, passage and rejoining of double-stranded DNA (according to Swiss-Prot)
- Protein family: topoisomerase gyrA/parC subunit family (according to Swiss-Prot)
- Paralogous protein(s): ParC
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: P05653
- KEGG entry: [3]
- E.C. number:
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Operon: gyrA PubMed
- Regulation:
- Regulatory mechanism:
- Additional information: subject to Clp-dependent proteolysis upon glucose starvation PubMed, GyrA is subject to Clp-dependent proteolysis upon glucose starvation PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Dagmar Klostermeier, Biozentrum Basel, Switzerland homepage
Your additional remarks
References
Markus G Rudolph, Dagmar Klostermeier
Mapping the spectrum of conformational states of the DNA- and C-gates in Bacillus subtilis gyrase.
J Mol Biol: 2013, 425(15);2632-40
[PubMed:23602808]
[WorldCat.org]
[DOI]
(I p)
Luise A K Kleine Borgmann, Hanna Hummel, Maximilian H Ulbrich, Peter L Graumann
SMC condensation centers in Bacillus subtilis are dynamic structures.
J Bacteriol: 2013, 195(10);2136-45
[PubMed:23475963]
[WorldCat.org]
[DOI]
(I p)
Nicole M Baker, Steven Weigand, Sarah Maar-Mathias, Alfonso Mondragón
Solution structures of DNA-bound gyrase.
Nucleic Acids Res: 2011, 39(2);755-66
[PubMed:20870749]
[WorldCat.org]
[DOI]
(I p)
Airat Gubaev, Manuel Hilbert, Dagmar Klostermeier
The DNA-gate of Bacillus subtilis gyrase is predominantly in the closed conformation during the DNA supercoiling reaction.
Proc Natl Acad Sci U S A: 2009, 106(32);13278-83
[PubMed:19666507]
[WorldCat.org]
[DOI]
(I p)
Ulf Gerth, Holger Kock, Ilja Kusters, Stephan Michalik, Robert L Switzer, Michael Hecker
Clp-dependent proteolysis down-regulates central metabolic pathways in glucose-starved Bacillus subtilis.
J Bacteriol: 2008, 190(1);321-31
[PubMed:17981983]
[WorldCat.org]
[DOI]
(I p)
Thomas Göttler, Dagmar Klostermeier
Dissection of the nucleotide cycle of B. subtilis DNA gyrase and its modulation by DNA.
J Mol Biol: 2007, 367(5);1392-404
[PubMed:17320901]
[WorldCat.org]
[DOI]
(P p)
Jean-Christophe Meile, Ling Juan Wu, S Dusko Ehrlich, Jeff Errington, Philippe Noirot
Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory.
Proteomics: 2006, 6(7);2135-46
[PubMed:16479537]
[WorldCat.org]
[DOI]
(P p)
W M Huang, J L Libbey, P van der Hoeven, S X Yu
Bipolar localization of Bacillus subtilis topoisomerase IV, an enzyme required for chromosome segregation.
Proc Natl Acad Sci U S A: 1998, 95(8);4652-7
[PubMed:9539793]
[WorldCat.org]
[DOI]
(P p)
N Ogasawara, S Moriya, H Yoshikawa
Structure and function of the region of the replication origin of the Bacillus subtilis chromosome. IV. Transcription of the oriC region and expression of DNA gyrase genes and other open reading frames.
Nucleic Acids Res: 1985, 13(7);2267-79
[PubMed:2987848]
[WorldCat.org]
[DOI]
(P p)