Crh
- Description: "Catabolite rpression HPr-like protein", Cofactor of the CcpA transcription factor
| Gene name | crh |
| Synonyms | yvcM |
| Essential | no |
| Product | catabolite repression HPr-like protein |
| Function | catabolite repression |
| MW, pI | 9,2 kDa, 4.70 |
| Gene length, protein length | 255 bp, 85 amino acids |
| Immediate neighbours | yvcL, yvcN |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 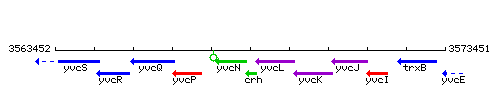
| |
Contents
The gene
Basic information
- Coordinates: 3568325 - 3568579
Phenotypes of a mutant
Database entries
- DBTBS entry:
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: PtsH,HPr family
- Paralogous protein(s): HPr
Extended information on the protein
- Kinetic information:
- Domains: HPr domain (1–85)
- Modification: phosphorylation at Ser46 by HPrK
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- Structure: NCBI, dimer NCBI, CcpA-Crh-DNA complex NCBI, dimeric phosphor-Crh NCBI
- Swiss prot entry: [2]
- KEGG entry: [3]
Additional information
Crh does not possess the phosphorylation site used for PTS phosphotransfer (His-15 in PtsH), it can only be phosphorylated on Ser-46
Expression and regulation
- Regulation: very weak stimuation of expression by citrate and succinate PubMed
- Regulatory mechanism:
- Additional information: Crh is weakly expressed. This results in part from a poorly conserved ribosomal binding site of the mRNA. PubMed
Biological materials
- Mutant: GP860 (aphA3), QB7097 (spc), available in Stülke lab
- Expression vector: pGP641 (in pGP380, for SPINE, expression in B. subtilis), pGP734 (in pGP380, for SPINE, expression in B. subtilis), available in Stülke lab
- lacZ fusion: see yvcI
- GFP fusion:
- Antibody: available in Stülke lab (not very good)
Labs working on this gene/protein
Boris Görke, University of Göttingen, Germany Homepage
Anne Galinier, University of Marseille, France
Your additional remarks
References
- Juy M, Penin F, Favier A (2003) Dimerization of Crh by reversible 3D domain swapping induces structural adjustments to its monomeric homologue Hpr. J Mol Biol. 332(4):767-76. PubMed
- Galinier A, Deutscher J, Martin-Verstraete I: Phosphorylation of either Crh or HPr mediates binding of CcpA to the Bacillus subtilis xyn cre and catabolite repression of the xyn operon. J Mol Biol 1999, 286:307-314. PubMed
- Galinier, A., Haiech, J., Kilhoffer, M.-C., Jaquinod, M., Stülke, J., Deutscher, J., & Martin-Verstraete, I. (1997) The Bacillus subtilis crh gene encodes a HPr-like protein involved in carbon catabolite repression. Proc. Natl. Acad. Sci. USA 94: 8439-8444. PubMed
- Görke, B., Fraysse, L. & Galinier, A. Drastic differences in Crh and HPr synthesis levels reflect their different impacts on catabolite repression in Bacillus subtilis. J. Bacteriol. 186, 2992-2995 (2004). PubMed
- Martin-Verstraete, I., Deutscher, J., and Galinier, A. (1999) Phosphorylation of HPr and Crh by HprK, early steps in the catabolite repression signalling pathway for the Bacillus subtilis levanase operon. J Bacteriol 181: 2966-2969. PubMed
- Pompeo et al. (2007) Interaction of GapA with HPr and its homologue, Crh: Novel levels of regulation of a key step of glycolysis in Bacillus subtilis? J Bacteriol 189, 1154-1157.PubMed
- Schumacher, M. A., Seidel, G., Hillen, W. & Brennan, R. G. Phosphoprotein Crh-Ser46-P displays altered binding to CcpA to effect carbon catabolite regulation. J. Biol. Chem. 281, 6793-6800 (2006). PubMed