BshB2
Revision as of 13:01, 13 May 2013 by 134.76.70.252 (talk)
- Description: N-acetylglucosamin-malate deacetylase, involved in bacillithiol synthesis
| Gene name | bshB2 |
| Synonyms | yojG |
| Essential | no |
| Product | N-acetylglucosamin-malate deacetylase |
| Function | biosynthesis of bacillithiol |
| Gene expression levels in SubtiExpress: bshB2 | |
| MW, pI | 16 kDa, 4.511 |
| Gene length, protein length | 426 bp, 142 aa |
| Immediate neighbours | rsbRC, yojF |
| Sequences | Protein DNA Advanced_DNA |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed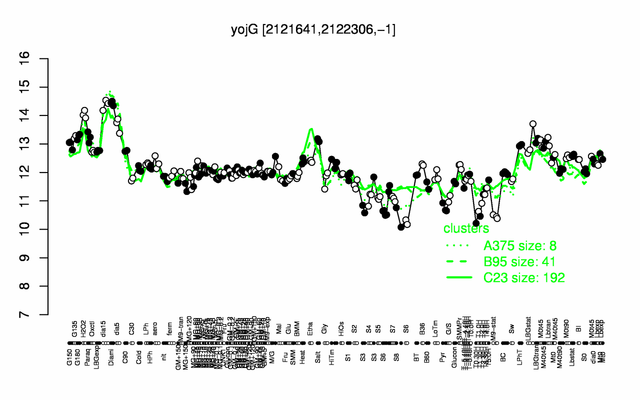
| |
Contents
Categories containing this gene/protein
miscellaneous metabolic pathways, resistance against oxidative and electrophile stress
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU19460
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s): BshB1
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: O31857
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: bshB2 null mutant and bshB1 / bshB2 double null mutant available in John Helmann lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Ahmed Gaballa, Gerald L Newton, Haike Antelmann, Derek Parsonage, Heather Upton, Mamta Rawat, Al Claiborne, Robert C Fahey, John D Helmann
Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli.
Proc Natl Acad Sci U S A: 2010, 107(14);6482-6
[PubMed:20308541]
[WorldCat.org]
[DOI]
(I p)
Gerald L Newton, Mamta Rawat, James J La Clair, Vishnu Karthik Jothivasan, Tanya Budiarto, Chris J Hamilton, Al Claiborne, John D Helmann, Robert C Fahey
Bacillithiol is an antioxidant thiol produced in Bacilli.
Nat Chem Biol: 2009, 5(9);625-7
[PubMed:19578333]
[WorldCat.org]
[DOI]
(I p)