MurG
- Description: UDP-N-acetylglucosamine-N-acetylmuramyl-(pentapeptide)pyrophosphoryl-undecaprenol N-acetylglucosamine transferase
| Gene name | murG |
| Synonyms | |
| Essential | yes PubMed |
| Product | UDP-N-acetylglucosamine-
N-acetylmuramyl-(pentapeptide)pyrophosphoryl- |
| Function | peptidoglycan precursor biosynthesis |
| Gene expression levels in SubtiExpress: murG | |
| Metabolic function and regulation of this protein in SubtiPathways: murG | |
| MW, pI | 39 kDa, 9.568 |
| Gene length, protein length | 1089 bp, 363 aa |
| Immediate neighbours | spoVE, murB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 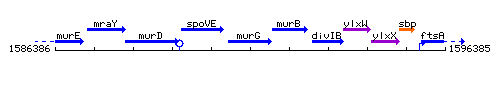
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
cell wall synthesis, biosynthesis of cell wall components, sporulation proteins, essential genes, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU15220
Phenotypes of a mutant
essential PubMed
Database entries
- BsubCyc: BSU15220
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- converts lipid I to lipid II by attaching GlcNAc to the bactoprenol carrier-conjugated UDP-N-acetylmuramic acid (UDP-MurNAc) pentapeptide molecule
- UDP-N-acetylglucosamine + Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)-diphosphoundecaprenol = UDP + GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)-diphosphoundecaprenol (according to Swiss-Prot)
- Protein family: MurG subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Localization:
- peripheral membrane protein PubMed
Database entries
- BsubCyc: BSU15220
- UniProt: P37585
- KEGG entry: [3]
- E.C. number: 2.4.1.227
Additional information
Expression and regulation
- Operon:
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- FLAG-tag construct:
- GP2002 murG-3xFLAG ermC (based on pGP1087), available in Jörg Stülke's lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References