SodA
- Description: superoxide dismutase, general stress protein
| Gene name | sodA |
| Synonyms | yqgD |
| Essential | no |
| Product | superoxide dismutase |
| Function | detoxification of oxygen radicals |
| MW, pI | 22 kDa, 5.203 |
| Gene length, protein length | 606 bp, 202 aa |
| Immediate neighbours | yqgE, yqgC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 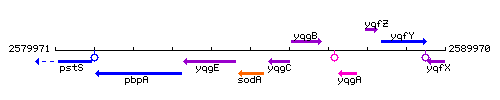
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag:
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 2 superoxide + 2 H+ = O2 + H2O2 (according to Swiss-Prot)
- Protein family: iron/manganese superoxide dismutase family (according to Swiss-Prot)
- Paralogous protein(s): SodF
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: phosphorylation on Thr-34 AND Thr-70 PubMed
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure: 2RCV
- Swiss prot entry: P54375
- KEGG entry: BSU25020
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
- Höper et al. (2005) Comprehensive Characterization of the Contribution of Individual SigB-Dependent General Stress Genes to Stress Resistance of Bacillus subtilis. J. Bact. 187: 2810-2826 PubMed
- Macek et al. (2007) The serine/ threonine/ tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6: 697-707 PubMed
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed