DegU
Revision as of 13:24, 8 December 2014 by Fabian Commichau (talk | contribs) (→Biological materials)
- Description: two-component response regulator, regulation of degradative enzyme expression, genetic competence, biofilm formation, capsule biosynthesis (together with SwrAA), non-phosphorylated DegU is required for swarming motility
| Gene name | degU |
| Synonyms | sacU, iep |
| Essential | no |
| Product | two-component response regulator |
| Function | regulation of degradative enzymes, genetic competence,
biofilm formation and capsule synthesis |
| Gene expression levels in SubtiExpress: degU | |
| Interactions involving this protein in SubtInteract: DegU | |
| Metabolic function and regulation of this protein in SubtiPathways: DegU | |
| MW, pI | 25 kDa, 5.602 |
| Gene length, protein length | 687 bp, 229 aa |
| Immediate neighbours | yviA, degS |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 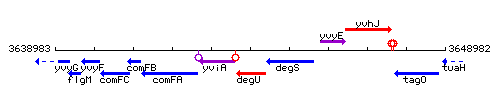
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed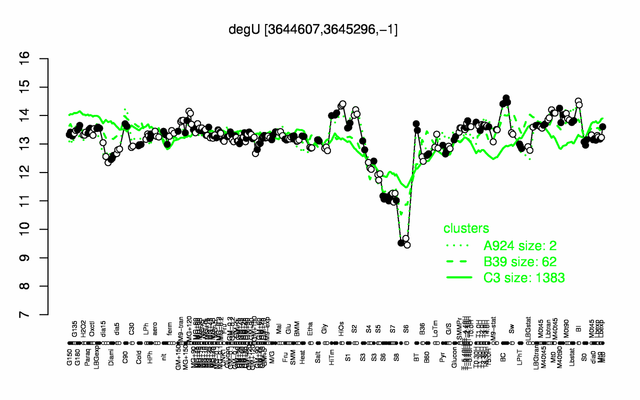
| |
Contents
Categories containing this gene/protein
biofilm formation, transcription factors and their control, phosphoproteins
This gene is a member of the following regulons
CcpA regulon, DegU regulon, SinR regulon, TnrA regulon
The DegU regulon
The gene
Basic information
- Locus tag: BSU35490
Phenotypes of a mutant
- defect in biofilm formation PubMed, this can be suppressed by DegU-independent expression of BslA PubMed
- the mutation suppresses the mucoid phenotype of motA or motB mutants due to reduced expression of the capB-capC-capA-capE operon PubMed
Database entries
- BsubCyc: BSU35490
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- transcription activator (see DegU regulon)
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU35490
- Structure:
- UniProt: P13800
- KEGG entry: [3]
- E.C. number:
Additional information
- DegU-P is degraded by ClpC-ClpP PubMed
- accumulation of DegU-P results in decreased transcription of the epsA-epsO and tapA-sipW-tasA operons due to increased levels of Spo0A-P PubMed
Expression and regulation
- Regulatory mechanism:
- Additional information:
- DegU-P is degraded by ClpC-ClpP PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 1627 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 2454 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 2251 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 1461 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 1432 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
The DegU regulon
Original Publications