LicH
- Description: 6-phospho-beta-glucosidase
| Gene name | licH |
| Synonyms | celD, celF |
| Essential | no |
| Product | 6-phospho-beta-glucosidase |
| Function | lichenan utilization |
| Gene expression levels in SubtiExpress: licH | |
| Metabolic function and regulation of this protein in SubtiPathways: licH | |
| MW, pI | 48 kDa, 5.183 |
| Gene length, protein length | 1326 bp, 442 aa |
| Immediate neighbours | ywaA, licA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 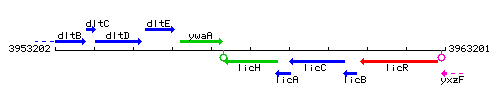
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed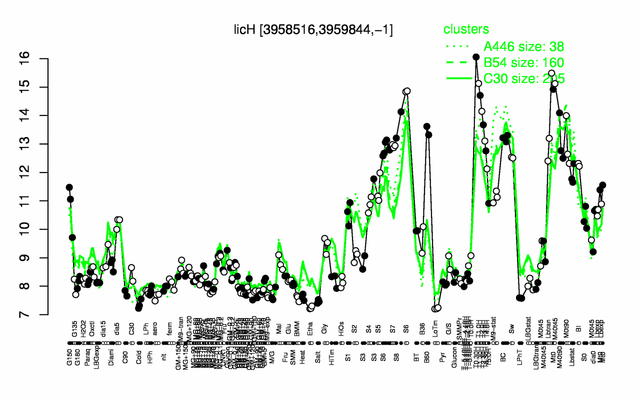
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources, sporulation proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU38560
Phenotypes of a mutant
Database entries
- BsubCyc: BSU38560
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 6-phospho-beta-D-glucosyl-(1,4)-D-glucose + H2O = D-glucose + D-glucose 6-phosphate (according to Swiss-Prot)
- Protein family: glycosyl hydrolase 4 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU38560
- Structure: 1S6Y (Geobacillus stearothermophilus)
- UniProt: P46320
- KEGG entry: [3]
- E.C. number: 3.2.1.86
Additional information
Expression and regulation
- Regulation:
- Additional information:
- number of protein molecules per cell (complex medium with amino acids, without glucose): 363 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References