Difference between revisions of "Eno"
(→References) |
|||
| Line 129: | Line 129: | ||
=References= | =References= | ||
| − | + | <pubmed> 17726680, 17218307, 12850135 , 19193632 , 11489127, 8021172, 17505547 </pubmed> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Revision as of 07:04, 2 June 2009
- Description: enolase, glycolytic/ gluconeogenic enzyme
| Gene name | eno |
| Synonyms | |
| Essential | yes |
| Product | enolase |
| Function | enzyme in glycolysis/ gluconeogenesis |
| MW, pI | 46,4 kDa, 4.49 |
| Gene length, protein length | 1290 bp, 430 amino acids |
| Immediate neighbours | pgm, yvgK |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 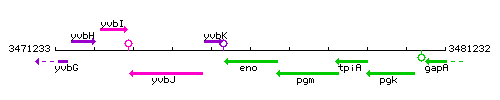
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Coordinates: 3475589 - 3476878
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 2-phospho-D-glycerate = phosphoenolpyruvate + H2O (according to Swiss-Prot) 2-phospho-D-glycerate = phosphoenolpyruvate + H(2)O
- Protein family: enolase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- substrate binding domain (366–369)
- Cofactor(s): magnesium ion
- Effectors of protein activity:
Database entries
- Structure:
- Swiss prot entry: P37869
- KEGG entry: BSU33900
- E.C. number: 4.2.1.11
Additional information
Expression and regulation
- Sigma factor: SigA
- Regulation: expression activated by glucose (3.3 fold) PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector: pGP563 (N-terminal His-tag, in pWH844), pGP93 (N-terminal Strep-tag, purification from B. subtilis, for SPINE, in pGP380), available in Stülke lab
- lacZ fusion:
- GFP fusion: pHT315-yfp-eno, available in Mijakovic lab
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody: available in Stülke lab
Labs working on this gene/protein
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References
Fabian M Commichau, Fabian M Rothe, Christina Herzberg, Eva Wagner, Daniel Hellwig, Martin Lehnik-Habrink, Elke Hammer, Uwe Völker, Jörg Stülke
Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing.
Mol Cell Proteomics: 2009, 8(6);1350-60
[PubMed:19193632]
[WorldCat.org]
[DOI]
(I p)
Christine Eymann, Dörte Becher, Jörg Bernhardt, Katrin Gronau, Anja Klutzny, Michael Hecker
Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis.
Proteomics: 2007, 7(19);3509-26
[PubMed:17726680]
[WorldCat.org]
[DOI]
(P p)
Laurent Jannière, Danielle Canceill, Catherine Suski, Sophie Kanga, Bérengère Dalmais, Roxane Lestini, Anne-Françoise Monnier, Jérôme Chapuis, Alexander Bolotin, Marina Titok, Emmanuelle Le Chatelier, S Dusko Ehrlich
Genetic evidence for a link between glycolysis and DNA replication.
PLoS One: 2007, 2(5);e447
[PubMed:17505547]
[WorldCat.org]
[DOI]
(I e)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
H Ludwig, G Homuth, M Schmalisch, F M Dyka, M Hecker, J Stülke
Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon.
Mol Microbiol: 2001, 41(2);409-22
[PubMed:11489127]
[WorldCat.org]
[DOI]
(P p)
M A Leyva-Vazquez, P Setlow
Cloning and nucleotide sequences of the genes encoding triose phosphate isomerase, phosphoglycerate mutase, and enolase from Bacillus subtilis.
J Bacteriol: 1994, 176(13);3903-10
[PubMed:8021172]
[WorldCat.org]
[DOI]
(P p)