Difference between revisions of "GyrA"
| Line 82: | Line 82: | ||
* '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P05653 P05653] | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P05653 P05653] | ||
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU00070] | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU00070 BSU00070] |
* '''E.C. number:''' | * '''E.C. number:''' | ||
Revision as of 23:03, 13 May 2009
- Description: DNA-Gyrase (subunit A)
| Gene name | gyrA |
| Synonyms | nalA |
| Essential | yes PubMed |
| Product | DNA-Gyrase (subunit A) |
| Function | DNA supercoiling, initation of replication cycle and DNA elongation |
| MW, pI | 91 kDa, 5.215 |
| Gene length, protein length | 2463 bp, 821 aa |
| Immediate neighbours | gyrB, rrnO-16S |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 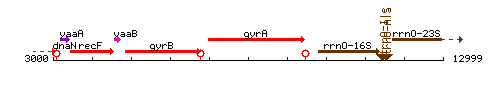
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: topoisomerase gyrA/parC subunit family (according to Swiss-Prot)
- Paralogous protein(s): ParC
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- Swiss prot entry: P05653
- KEGG entry: BSU00070
- E.C. number:
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Operon: gyrA PubMed
- Regulation:
- Regulatory mechanism:
- Additional information: subject to Clp-dependent proteolysis upon glucose starvation PubMed, GyrA is subject to Clp-dependent proteolysis upon glucose starvation PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Dagmar Klostermeier, Biozentrum Basel, Switzerland homepage
Your additional remarks
References
- Gerth et al. (2008) Clp-dependent proteolysis down-regulates central metabolic pathways in glucose-starved Bacillus subtilis. J Bacteriol 190:321-331 PubMed
- Meile et al. (2006) Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory Proteomics 6: 2135-2146. PubMed
- Huang et al. (1998) Bipolar localization of Bacillus subtilis topoisomerase IV, an enzyme required for chromosome segregation. Proc. Natl. Acad. Sci. USA 95: 4652-4657. PubMed
- Gerth et al. (2008) Clp-dependent proteolysis down-regulates central metabolic pathways in glucose-starved Bacillus subtilis. J Bacteriol 190:321-331 PubMed
- Ogasawara et al. (1985) Structure and function of the region of the replication origin of the Bacillus subtilis chromosome. IV. Transcription of the oriC region and expression of DNA gyrase genes and other open reading frames.Nucl. Acids Res. 11: 2267-2279. PubMed